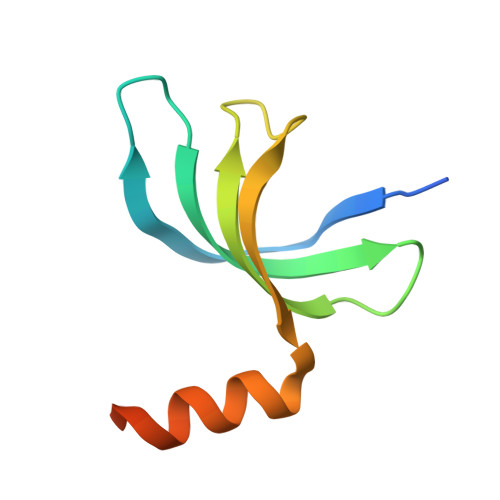
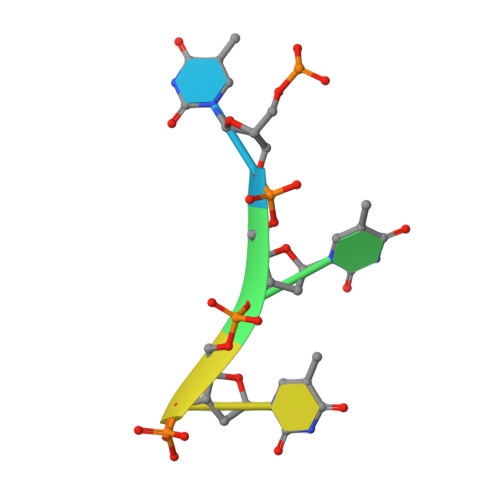
Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Li, S., Lu, G., Fang, X., Ramelot, T.A., Kennedy, M.A., Zhou, X., Gong, P., Zhang, X., Liu, M., Zhu, J., Yang, Y.(2020) Nucleic Acids Res 48: 432-444
- PubMed: 31713614
- DOI: https://doi.org/10.1093/nar/gkz1045
- Primary Citation of Related Structures:
5ZKL, 5ZKM, 6JIP, 6JIQ - PubMed Abstract:
SP_0782 from Streptococcus pneumoniae is a dimeric protein that potentially binds with single-stranded DNA (ssDNA) in a manner similar to human PC4, the prototype of PC4-like proteins, which plays roles in transcription and maintenance of genome stability. In a previous NMR study, SP_0782 exhibited an ssDNA-binding property different from YdbC, a prokaryotic PC4-like protein from Lactococcus lactis, but the underlying mechanism remains unclear. Here, we show that although SP_0782 adopts an overall fold similar to those of PC4 and YdbC, the ssDNA length occupied by SP_0782 is shorter than those occupied by PC4 and YdbC. SP_0782 exhibits varied binding patterns for different lengths of ssDNA, and tends to form large complexes with ssDNA in a potential high-density binding manner. The structures of SP_0782 complexed with different ssDNAs reveal that the varied binding patterns are associated with distinct capture of nucleotides in two major DNA-binding regions of SP_0782. Moreover, a comparison of known structures of PC4-like proteins complexed with ssDNA reveals a divergence in the binding interface between prokaryotic and eukaryotic PC4-like proteins. This study provides insights into the ssDNA-binding mechanism of PC4-like proteins, and benefits further study regarding the biological function of SP_0782, probably in DNA protection and natural transformation.
Organizational Affiliation:
State Key Laboratory of Magnetic Resonance and Atomic Molecular Physics, Key Laboratory of Magnetic Resonance in Biological Systems, National Center for Magnetic Resonance in Wuhan, Wuhan Institute of Physics and Mathematics, Chinese Academy of Sciences, Wuhan National Laboratory for Optoelectronics, Wuhan 430071, China.