Cyclic Peptidic Mimetics of Apollo Peptides Targeting Telomeric Repeat Binding Factor 2 (TRF2) and Apollo Interaction.
Chen, X., Liu, L., Chen, Y., Yang, Y., Yang, C.Y., Guo, T., Lei, M., Sun, H., Wang, S.(2018) ACS Med Chem Lett 9: 507-511
- PubMed: 29795768
- DOI: https://doi.org/10.1021/acsmedchemlett.8b00152
- Primary Citation of Related Structures:
6J67 - PubMed Abstract:
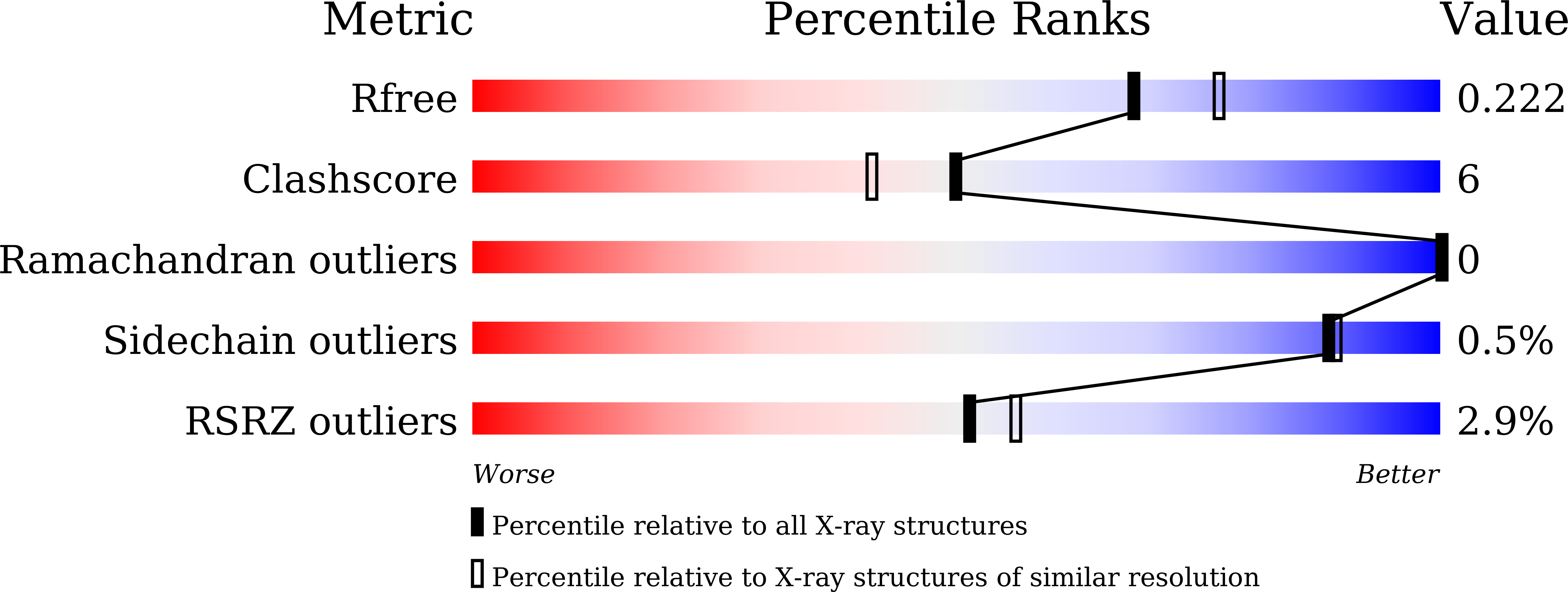
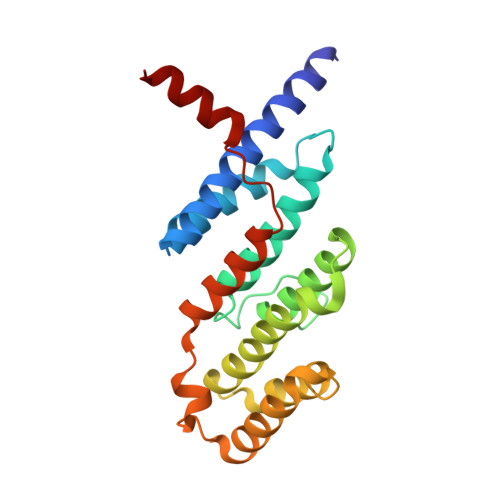
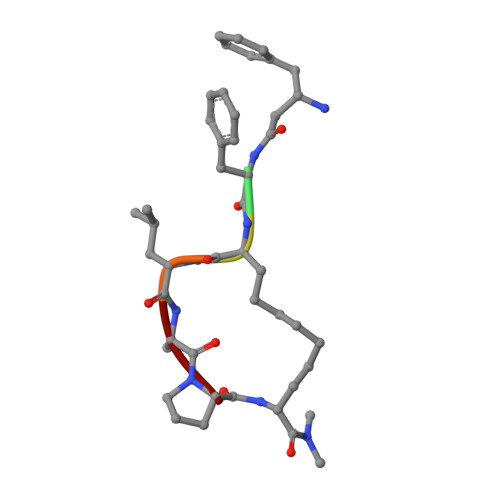
Telomeric repeat binding factor 2 (TRF2) is a telomere-associated protein that plays an important role in the formation of the 3' single strand DNA overhang and the "T loop", two structures critical for the stability of the telomeres. Apollo is a 5'-exonuclease recruited by TRF2 to the telomere and contributes to the formation of the 3' single strand DNA overhang. Knocking down of Apollo can induce DNA damage response similar to that caused by the knocking down of TRF2. In this Letter, we report the design and synthesis of a class of cyclic peptidic mimetics of the TRFH binding motif of Apollo (Apollo TBM ). We found conformational control of the C terminal residues of Apollo TBM can effectively improve the binding affinity. We have obtained a crystal structure of a cyclic peptidic Apollo peptide mimetic ( 34 ) complexed with TRF2, which provides valuable guidance to the future design of TRF2 inhibitors.
Organizational Affiliation:
Jiangsu Key Laboratory of Drug Design and Optimization, Department of Medicinal Chemistry, China Pharmaceutical University, Nanjing 210009, China.