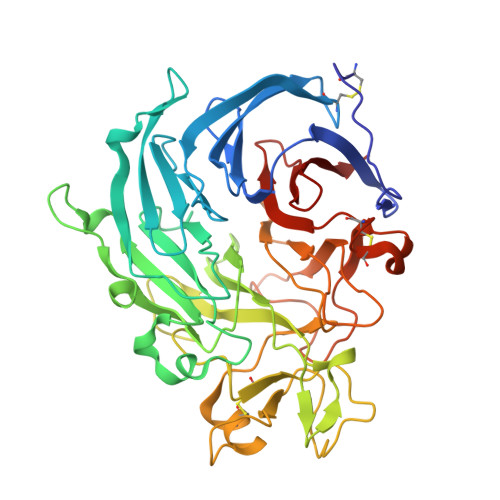
The functional and structural characterization ofTrichoderma reeseidehydrogenase belonging to the PQQ dependent family of Carbohydrate-Active Enzymes Family AA12.
Turbe-Doan, A., Record, E., Lombard, V., Kumar, R., Levasseur, A., Henrissat, B., Garron, M.L.(2019) Appl Environ Microbiol
- PubMed: 31604773
- DOI: https://doi.org/10.1128/AEM.00964-19
- Primary Citation of Related Structures:
6H7T, 6I1Q, 6I1T - PubMed Abstract:
Pyrroloquinoline quinone (PQQ) is an ortho -quinone cofactor of several prokaryotic oxidases. Widely available in the diet and necessary for the correct growth of mice, PQQ has been suspected to be a vitamin for eukaryotes. However, no PQQ-dependent eukaryotic enzyme had been identified to use the PQQ until 2014, when a basidiomycete enzyme catalyzing saccharide dehydrogenation using PQQ as a cofactor was characterized and served to define auxiliary activity family 12 (AA12). Here we report the biochemical characterization of the AA12 enzyme encoded by the genome of the ascomycete Trichoderma reesei ( Tr AA12). Surprisingly, only weak activity against uncommon carbohydrates like l-fucose or d-arabinose was measured. The three-dimensional structure of Tr AA12 reveals important similarities with bacterial soluble glucose dehydrogenases (sGDH). The enzymatic characterization and the structure solved in the presence of calcium confirm the importance of this ion in catalysis, as observed for sGDH. The structural characterization of Tr AA12 was completed by modeling PQQ and l-fucose in the enzyme active site. Based on these results, the AA12 family of enzymes is likely to have a catalytic mechanism close to that of bacterial sGDH. IMPORTANCE Pyrroloquinoline quinone (PQQ) is an important cofactor synthesized by prokaryotes and involved in enzymatic alcohol and sugar oxidation. In eukaryotes, the benefit of PQQ as a vitamin has been suggested but never proved. Recently, the first eukaryotic enzyme using PQQ was characterized in the basidiomycete Coprinopsis cinerea , demonstrating that fungi are able to use PQQ as an enzyme cofactor. This discovery led to the classification of the fungal PQQ-dependent enzymes in auxiliary activity family 12 (AA12) of the Carbohydrate-Active Enzymes (CAZy) database (www.cazy.org) classification. In the present paper, we report on the characterization of the ascomycete AA12 enzyme from Trichoderma reesei ( Tr AA12). Our enzymatic and phylogenetic results show divergence with the only other member of the family characterized, that from the basidiomycete Coprinopsis cinerea The crystallographic structure of Tr AA12 shows similarities to the global active-site architecture of bacterial glucose dehydrogenases, suggesting a common evolution between the two families.
Organizational Affiliation:
INRA, UMR1163, Biodiversité et Biotechnologie Fongiques, Aix-Marseille Université, Marseille, France.