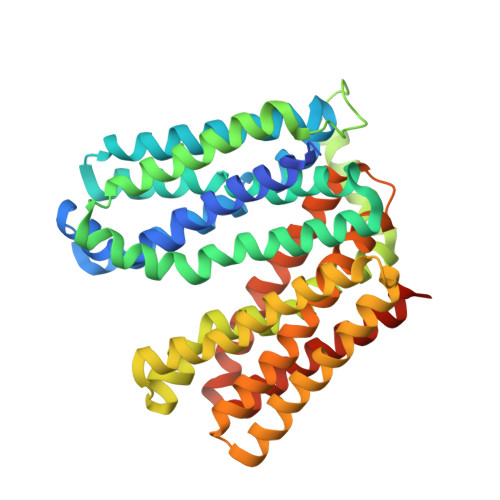
Outward open conformation of a Major Facilitator Superfamily multidrug/H+antiporter provides insights into switching mechanism.
Nagarathinam, K., Nakada-Nakura, Y., Parthier, C., Terada, T., Juge, N., Jaenecke, F., Liu, K., Hotta, Y., Miyaji, T., Omote, H., Iwata, S., Nomura, N., Stubbs, M.T., Tanabe, M.(2018) Nat Commun 9: 4005-4005
- PubMed: 30275448
- DOI: https://doi.org/10.1038/s41467-018-06306-x
- Primary Citation of Related Structures:
6GV1 - PubMed Abstract:
Multidrug resistance (MDR) poses a major challenge to medicine. A principle cause of MDR is through active efflux by MDR transporters situated in the bacterial membrane. Here we present the crystal structure of the major facilitator superfamily (MFS) drug/H + antiporter MdfA from Escherichia coli in an outward open conformation. Comparison with the inward facing (drug binding) state shows that, in addition to the expected change in relative orientations of the N- and C-terminal lobes of the antiporter, the conformation of TM5 is kinked and twisted. In vitro reconstitution experiments demonstrate the importance of selected residues for transport and molecular dynamics simulations are used to gain insights into antiporter switching. With the availability of structures of alternative conformational states, we anticipate that MdfA will serve as a model system for understanding drug efflux in MFS MDR antiporters.
Organizational Affiliation:
ZIK HALOmem, Kurt-Mothes-Straße 3, D-06120, Halle/Saale, Germany.