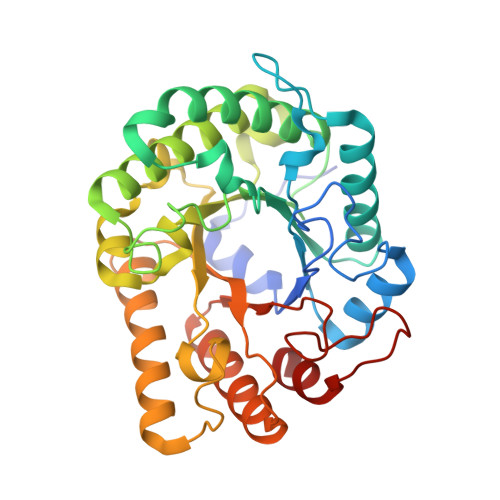
The laterally acquired GH5ZgEngAGH5_4from the marine bacteriumZobellia galactanivoransis dedicated to hemicellulose hydrolysis.
Dorival, J., Ruppert, S., Gunnoo, M., Orlowski, A., Chapelais-Baron, M., Dabin, J., Labourel, A., Thompson, D., Michel, G., Czjzek, M., Genicot, S.(2018) Biochem J 475: 3609-3628
- PubMed: 30341165
- DOI: https://doi.org/10.1042/BCJ20180486
- Primary Citation of Related Structures:
6GL0, 6GL2 - PubMed Abstract:
Cell walls of marine macroalgae are composed of diverse polysaccharides that provide abundant carbon sources for marine heterotrophic bacteria. Among them, Zobellia galactanivorans is considered as a model for studying algae-bacteria interactions. The degradation of typical algal polysaccharides, such as agars or alginate, has been intensively studied in this model bacterium, but the catabolism of plant-like polysaccharides is essentially uncharacterized. Here, we identify a polysaccharide utilization locus in the genome of Z. galactanivorans , induced by laminarin (β-1,3-glucans), and containing a putative GH5 subfamily 4 (GH5_4) enzyme, currently annotated as a endoglucanase ( Zg EngA GH5_4 ). A phylogenetic analysis indicates that Zg EngA GH5_4 was laterally acquired from an ancestral Actinobacteria We performed the biochemical and structural characterization of Zg EngA GH5_4 and demonstrated that this GH5 is, in fact, an endo-β-glucanase, most active on mixed-linked glucan (MLG). Although Zg EngA GH5_4 and GH16 lichenases both hydrolyze MLG, these two types of enzymes release different series of oligosaccharides. Structural analyses of Zg EngA GH5_4 reveal that all the amino acid residues involved in the catalytic triad and in the negative glucose-binding subsites are conserved, when compared with the closest relative, the cellulase EngD from Clostridium cellulovorans , and some other GH5s. In contrast, the positive glucose-binding subsites of Zg EngA GH5_4 are different and this could explain the preference for MLG, with respect to cellulose or laminarin. Molecular dynamics computer simulations using different hexaoses reveal that the specificity for MLG occurs through the +1 and +2 subsites of the binding pocket that display the most important differences when compared with the structures of other GH5_4 enzymes.
Organizational Affiliation:
Sorbonne Université, CNRS, Integrative Biology of Marine Models (LBI2M), Station Biologique de Roscoff (SBR), 29680 Roscoff, France.