Crystal structure of mammalian Rev7 in complex with Champ1 - CAPS molecule
Huber, F., Tropia, L., Emamzadah, S., Halazonetis, T.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Mitotic spindle assembly checkpoint protein MAD2B | 211 | Mus musculus | Mutation(s): 5 Gene Names: Mad2l2, Mad2b, Rev7 | 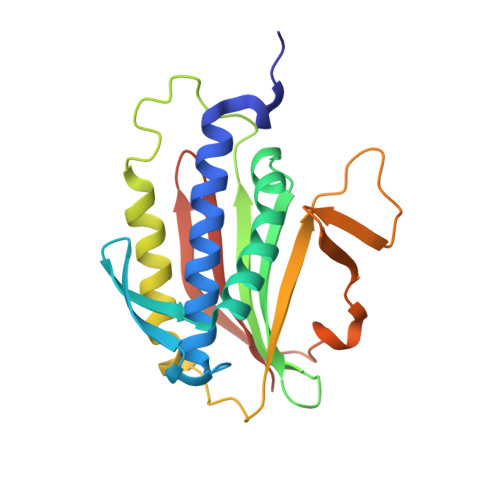 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9D752 (Mus musculus) Explore Q9D752 Go to UniProtKB: Q9D752 | |||||
IMPC: MGI:1919140 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9D752 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Chromosome alignment-maintaining phosphoprotein 1 | 29 | Homo sapiens | Mutation(s): 0 Gene Names: CHAMP1, C13orf8, CAMP, CHAMP, KIAA1802, ZNF828 | 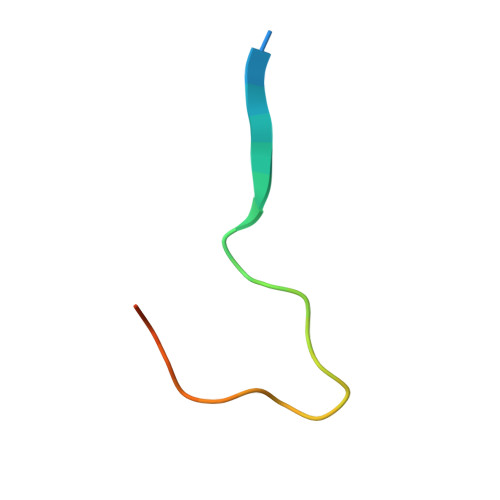 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q96JM3 (Homo sapiens) Explore Q96JM3 Go to UniProtKB: Q96JM3 | |||||
PHAROS: Q96JM3 GTEx: ENSG00000198824 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q96JM3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CXS Query on CXS | C [auth A] | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID C9 H19 N O3 S PJWWRFATQTVXHA-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 47.26 | α = 90 |
| b = 51.62 | β = 90 |
| c = 126 | γ = 90 |
| Software Name | Purpose |
|---|---|
| CNS | refinement |
| iMOSFLM | data reduction |
| SCALA | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Swiss National Science Foundation | Switzerland | 160322 |