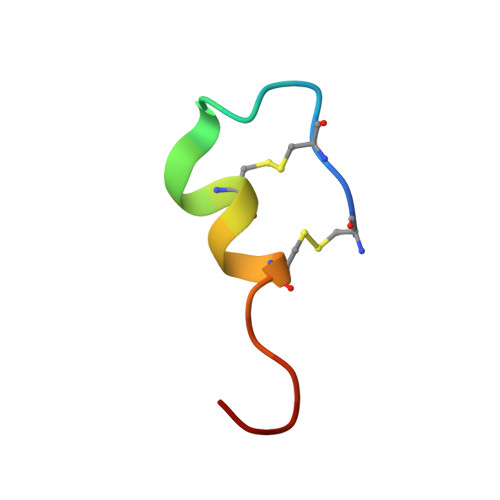
The X-ray crystal structure of human endothelin 1, a polypeptide hormone regulator of blood pressure.
McPherson, A., Larson, S.B.(2019) Acta Crystallogr F Struct Biol Commun 75: 47-53
- PubMed: 30605125
- DOI: https://doi.org/10.1107/S2053230X18016011
- Primary Citation of Related Structures:
6DK5 - PubMed Abstract:
Human endothelin is a 21-amino-acid polypeptide, constrained by two intra-chain disulfide bridges, that is made by endothelial cells. It is the most potent vasoconstrictor in the body and is crucially important in the regulation of blood pressure. It plays a major role in a host of medical conditions, including hypertension, diabetes, stroke and cancer. Endothelin was crystallized 28 years ago in the putative space group P6 1 22, but the structure was never successfully solved by X-ray diffraction. Using X-ray diffraction data from 1992, the structure has now been solved. Assuming a unit cell belonging to space group P6 1 and a twin fraction of 0.28, a solution emerged with two, almost identical, closely associated molecules in the asymmetric unit. Although the data extended to beyond 1.8 Å resolution, a model containing 25 waters was refined to 1.85 Å resolution with an R of 0.216 and an R free of 0.284. The disulfide-constrained `core' of the molecule, amino-acid residues 1-15, has a main-chain conformation that is essentially the same as endothelin when bound to its receptor, but many side-chain rotamers are different. The carboxy-terminal `tail' comprising amino-acid residues 16-21 is extended as when receptor-bound, but it exhibits a different conformation with respect to the `core'. The dimer that comprises the asymmetric unit is maintained almost exclusively by hydrophobic interactions and may be stable in an aqueous medium.
Organizational Affiliation:
Molecular Biology and Biochemistry, University of California Irvine, McGaugh Hall, Irvine, CA 92697-3900, USA.