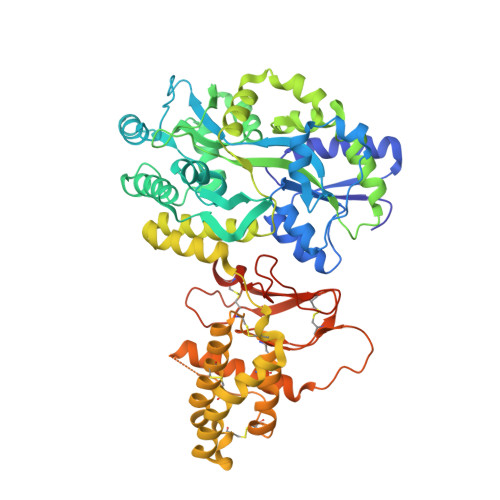
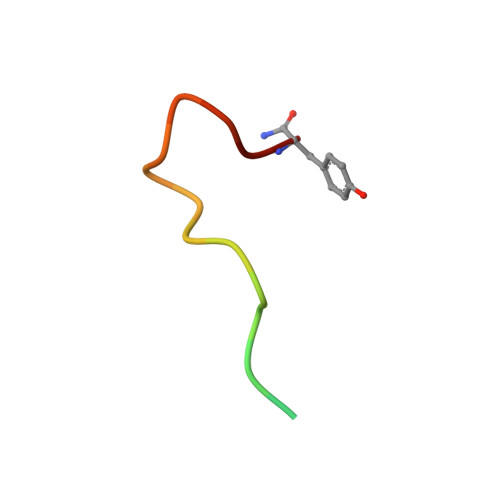
Structure-function analyses reveal a triple beta-turn receptor-bound conformation of adrenomedullin 2/intermedin and enable peptide antagonist design.
Roehrkasse, A.M., Booe, J.M., Lee, S.M., Warner, M.L., Pioszak, A.A.(2018) J Biol Chem 293: 15840-15854
- PubMed: 30139742
- DOI: https://doi.org/10.1074/jbc.RA118.005062
- Primary Citation of Related Structures:
6D1U - PubMed Abstract:
The cardioprotective vasodilator peptide adrenomedullin 2/intermedin (AM2/IMD) and the related adrenomedullin (AM) and calcitonin gene-related peptide (CGRP) signal through three heterodimeric receptors comprising the calcitonin receptor-like class B G protein-coupled receptor (CLR) and a variable receptor activity-modifying protein (RAMP1, -2, or -3) that determines ligand selectivity. The CGRP receptor (RAMP1:CLR) favors CGRP binding, whereas the AM 1 (RAMP2:CLR) and AM 2 (RAMP3:CLR) receptors favor AM binding. How AM2/IMD binds the receptors and how RAMPs modulate its binding is unknown. Here, we show that AM2/IMD binds the three purified RAMP-CLR extracellular domain (ECD) complexes with a selectivity profile that is distinct from those of CGRP and AM. AM2/IMD bound all three ECD complexes but preferred the CGRP and AM 2 receptor complexes. A 2.05 Å resolution crystal structure of an AM2/IMD antagonist fragment-bound RAMP1-CLR ECD complex revealed that AM2/IMD binds the complex through a unique triple β-turn conformation that was confirmed by peptide and receptor mutagenesis. Comparisons of the receptor-bound conformations of AM2/IMD, AM, and a high-affinity CGRP analog revealed differences that may have implications for biased signaling. Guided by the structure, enhanced-affinity AM2/IMD antagonist variants were developed, including one that discriminates the AM 1 and AM 2 receptors with ∼40-fold difference in affinities and one stabilized by an intramolecular disulfide bond. These results reveal differences in how the three peptides engage the receptors, inform development of AM2/IMD-based pharmacological tools and therapeutics, and provide insights into RAMP modulation of receptor pharmacology.
Organizational Affiliation:
From the Department of Biochemistry and Molecular Biology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma 73104.