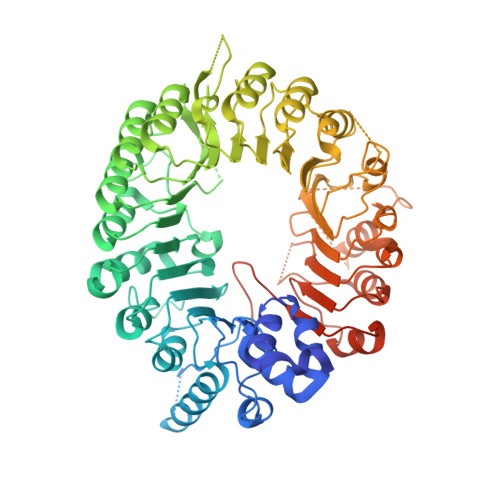
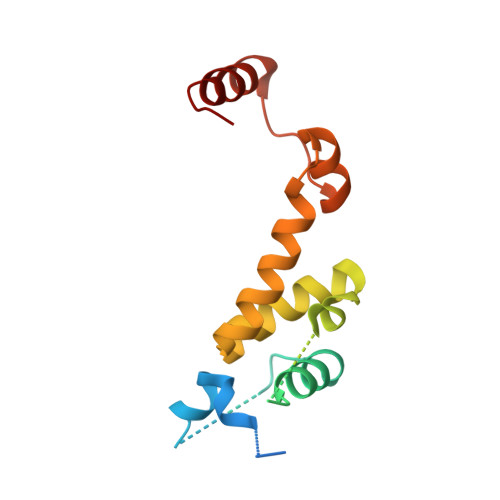
Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling.
Shabek, N., Ticchiarelli, F., Mao, H., Hinds, T.R., Leyser, O., Zheng, N.(2018) Nature 563: 652-656
- PubMed: 30464344
- DOI: https://doi.org/10.1038/s41586-018-0743-5
- Primary Citation of Related Structures:
6BRO, 6BRP, 6BRQ, 6BRT - PubMed Abstract:
The strigolactones, a class of plant hormones, regulate many aspects of plant physiology. In the inhibition of shoot branching, the α/β hydrolase D14-which metabolizes strigolactone-interacts with the F-box protein D3 to ubiquitinate and degrade the transcription repressor D53. Despite the fact that multiple modes of interaction between D14 and strigolactone have recently been determined, how the hydrolase functions with D3 to mediate hormone-dependent D53 ubiquitination remains unknown. Here we show that D3 has a C-terminal α-helix that can switch between two conformational states. The engaged form of this α-helix facilitates the binding of D3 and D14 with a hydrolysed strigolactone intermediate, whereas the dislodged form can recognize unmodified D14 in an open conformation and inhibits its enzymatic activity. The D3 C-terminal α-helix enables D14 to recruit D53 in a strigolactone-dependent manner, which in turn activates the hydrolase. By revealing the structural plasticity of the SCF D3-D14 ubiquitin ligase, our results suggest a mechanism by which the E3 coordinates strigolactone signalling and metabolism.
Organizational Affiliation:
Department of Pharmacology, University of Washington, Seattle, WA, USA.