Uncovering a novel molecular mechanism for scavenging sialic acids in bacteria.
Bell, A., Severi, E., Lee, M., Monaco, S., Latousakis, D., Angulo, J., Thomas, G.H., Naismith, J.H., Juge, N.(2020) J Biol Chem 295: 13724-13736
- PubMed: 32669363
- DOI: https://doi.org/10.1074/jbc.RA120.014454
- Primary Citation of Related Structures:
6Z3B, 6Z3C - PubMed Abstract:
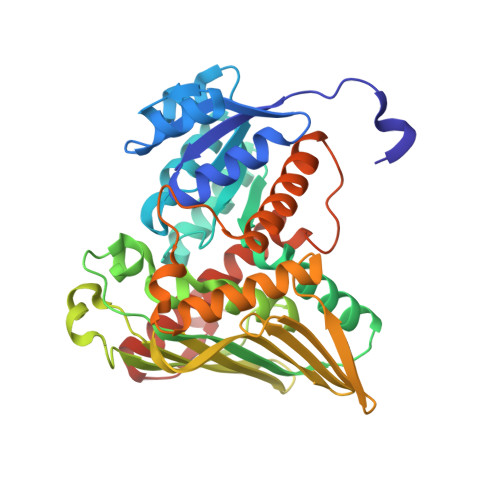
The human gut symbiont Ruminococcus gnavus scavenges host-derived N -acetylneuraminic acid (Neu5Ac) from mucins by converting it to 2,7-anhydro-Neu5Ac. We previously showed that 2,7-anhydro-Neu5Ac is transported into R. gnavus ATCC 29149 before being converted back to Neu5Ac for further metabolic processing. However, the molecular mechanism leading to the conversion of 2,7-anhydro-Neu5Ac to Neu5Ac remained elusive. Using 1D and 2D NMR, we elucidated the multistep enzymatic mechanism of the oxidoreductase ( Rg NanOx) that leads to the reversible conversion of 2,7-anhydro-Neu5Ac to Neu5Ac through formation of a 4-keto-2-deoxy-2,3-dehydro- N -acetylneuraminic acid intermediate and NAD + regeneration. The crystal structure of Rg NanOx in complex with the NAD + cofactor showed a protein dimer with a Rossman fold. Guided by the Rg NanOx structure, we identified catalytic residues by site-directed mutagenesis. Bioinformatics analyses revealed the presence of Rg NanOx homologues across Gram-negative and Gram-positive bacterial species and co-occurrence with sialic acid transporters. We showed by electrospray ionization spray MS that the Escherichia coli homologue YjhC displayed activity against 2,7-anhydro-Neu5Ac and that E. coli could catabolize 2,7-anhydro-Neu5Ac. Differential scanning fluorimetry analyses confirmed the binding of YjhC to the substrates 2,7-anhydro-Neu5Ac and Neu5Ac, as well as to co-factors NAD and NADH. Finally, using E. coli mutants and complementation growth assays, we demonstrated that 2,7-anhydro-Neu5Ac catabolism in E. coli depended on YjhC and on the predicted sialic acid transporter YjhB. These results revealed the molecular mechanisms of 2,7-anhydro-Neu5Ac catabolism across bacterial species and a novel sialic acid transport and catabolism pathway in E. coli .
Organizational Affiliation:
Gut Microbes and Health Institute Strategic Programme, Quadram Institute Bioscience, Norwich, United Kingdom.