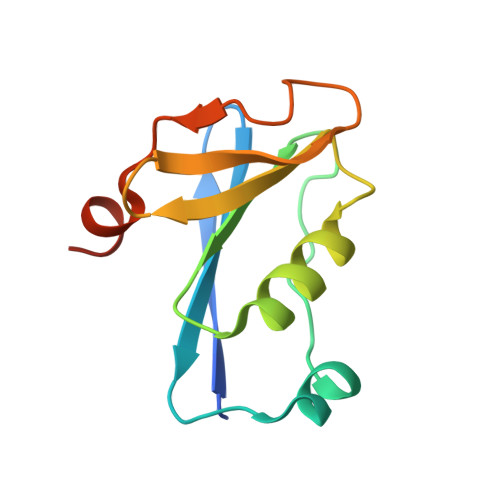
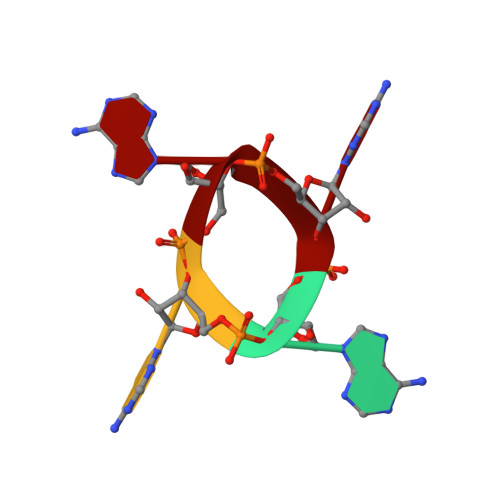
Tetramerisation of the CRISPR ring nuclease Crn3/Csx3 facilitates cyclic oligoadenylate cleavage.
Athukoralage, J.S., McQuarrie, S., Gruschow, S., Graham, S., Gloster, T.M., White, M.F.(2020) Elife 9
- PubMed: 32597755
- DOI: https://doi.org/10.7554/eLife.57627
- Primary Citation of Related Structures:
6YUD - PubMed Abstract:
Type III CRISPR systems detect foreign RNA and activate the cyclase domain of the Cas10 subunit, generating cyclic oligoadenylate (cOA) molecules that act as a second messenger to signal infection, activating nucleases that degrade the nucleic acid of both invader and host. This can lead to dormancy or cell death; to avoid this, cells need a way to remove cOA from the cell once a viral infection has been defeated. Enzymes specialised for this task are known as ring nucleases, but are limited in their distribution. Here, we demonstrate that the widespread CRISPR associated protein Csx3, previously described as an RNA deadenylase, is a ring nuclease that rapidly degrades cyclic tetra-adenylate (cA 4 ). The enzyme has an unusual cooperative reaction mechanism involving an active site that spans the interface between two dimers, sandwiching the cA 4 substrate. We propose the name Crn3 (CRISPR associated ring nuclease 3) for the Csx3 family.
Organizational Affiliation:
Biomedical Sciences Research Complex, School of Biology, University of St Andrews, St Andrews, United Kingdom.