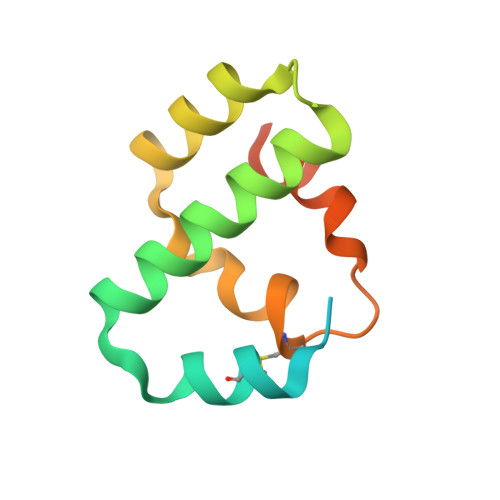
Crystal structure of bacteriophage T4 Spackle as determined by native SAD phasing.
Shi, K., Kurniawan, F., Banerjee, S., Moeller, N.H., Aihara, H.(2020) Acta Crystallogr D Struct Biol 76: 899-904
- PubMed: 32876065
- DOI: https://doi.org/10.1107/S2059798320010979
- Primary Citation of Related Structures:
6X6O - PubMed Abstract:
The crystal structure of a bacteriophage T4 early gene product, Spackle, was determined by native sulfur single-wavelength anomalous diffraction (SAD) phasing using synchrotron radiation and was refined to 1.52 Å resolution. The structure shows that Spackle consists of a bundle of five α-helices, forming a relatively flat disc-like overall shape. Although Spackle forms a dimer in the crystal, size-exclusion chromatography with multi-angle light scattering shows that it is monomeric in solution. Mass spectrometry confirms that purified mature Spackle lacks the amino-terminal signal peptide and contains an intramolecular disulfide bond, consistent with its proposed role in the periplasm of T4 phage-infected Escherichia coli cells. The surface electrostatic potential of Spackle shows a strikingly bipolar charge distribution, suggesting a possible mode of membrane association and inhibition of the tail lysozyme activity in T4 bacteriophage superinfection exclusion.
Organizational Affiliation:
Department of Biochemistry, Molecular Biology and Biophysics, University of Minnesota, Minneapolis, MN 55455, USA.