Crystallographic Characterization of the Carbonylated A-Cluster in Carbon Monoxide Dehydrogenase/Acetyl-CoA Synthase
Cohen, S.E., Can, M., Wittenborn, E.C., Hendrickson, R.A., Ragsdale, S.W., Drennan, C.L.(2020) ACS Catal 10: 9741-9746
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2020) ACS Catal 10: 9741-9746
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Carbon monoxide dehydrogenase/acetyl-CoA synthase subunit beta | 674 | Moorella thermoacetica | Mutation(s): 0 EC: 1.2.7.4 | 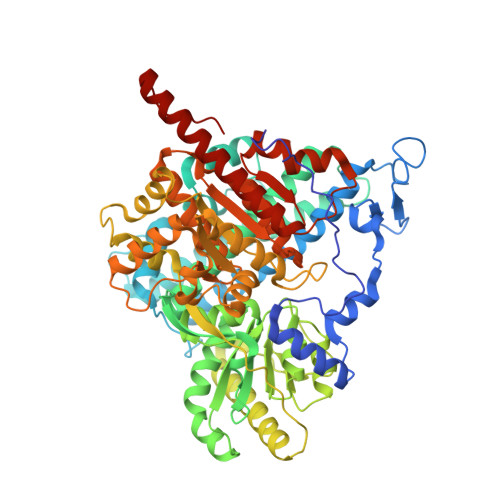 | |
UniProt | |||||
Find proteins for P27989 (Moorella thermoacetica) Explore P27989 Go to UniProtKB: P27989 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P27989 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Carbon monoxide dehydrogenase/acetyl-CoA synthase subunit alpha | E [auth M], F [auth N], G [auth O], H [auth P] | 729 | Moorella thermoacetica | Mutation(s): 0 EC: 2.3.1.169 | 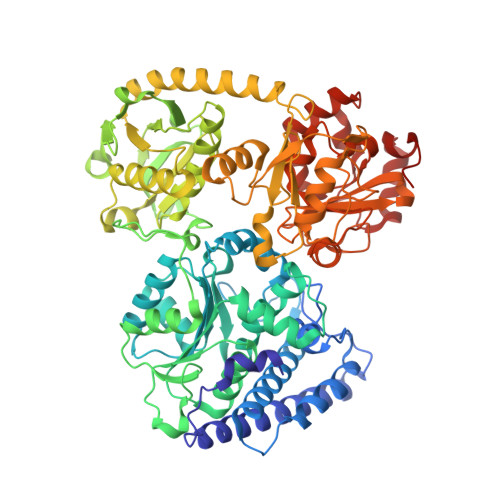 |
UniProt | |||||
Find proteins for P27988 (Moorella thermoacetica) Explore P27988 Go to UniProtKB: P27988 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P27988 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| XCC Query on XCC | K [auth A], N [auth B], Q [auth C], T [auth D] | FE(4)-NI(1)-S(4) CLUSTER Fe4 Ni S4 QGLWBXDZIHZONR-UHFFFAOYSA-N |  | ||
| SF4 (Subject of Investigation/LOI) Query on SF4 | DA [auth O] GA [auth P] I [auth A] J [auth A] M [auth B] | IRON/SULFUR CLUSTER Fe4 S4 LJBDFODJNLIPKO-UHFFFAOYSA-N |  | ||
| PG5 Query on PG5 | R [auth C] | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE C8 H18 O4 YFNKIDBQEZZDLK-UHFFFAOYSA-N |  | ||
| PGE Query on PGE | L [auth A] | TRIETHYLENE GLYCOL C6 H14 O4 ZIBGPFATKBEMQZ-UHFFFAOYSA-N |  | ||
| NI (Subject of Investigation/LOI) Query on NI | AA [auth N] EA [auth O] FA [auth O] HA [auth P] IA [auth P] | NICKEL (II) ION Ni VEQPNABPJHWNSG-UHFFFAOYSA-N |  | ||
| CMO (Subject of Investigation/LOI) Query on CMO | BA [auth N], JA [auth P], X [auth M] | CARBON MONOXIDE C O UGFAIRIUMAVXCW-UHFFFAOYSA-N |  | ||
| MG Query on MG | CA [auth N] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 99.729 | α = 101.818 |
| b = 137.383 | β = 109.077 |
| c = 141.867 | γ = 103.657 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM126982 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM008334 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM124165 |
| Department of Energy (DOE, United States) | United States | DE-AC02-06CH11357 |