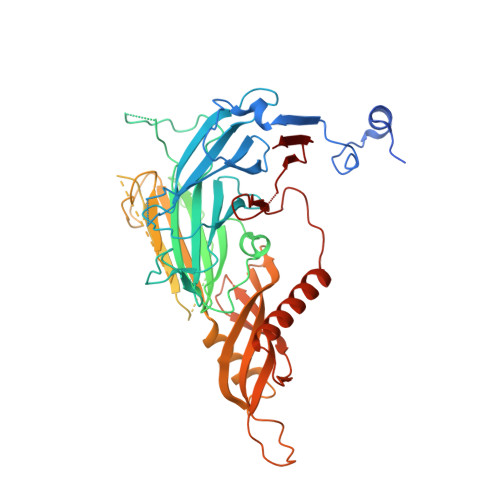
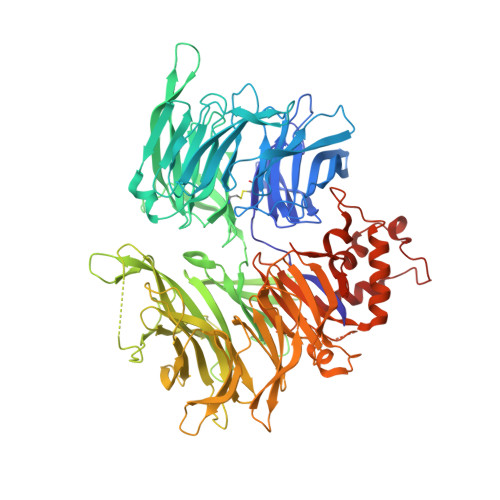
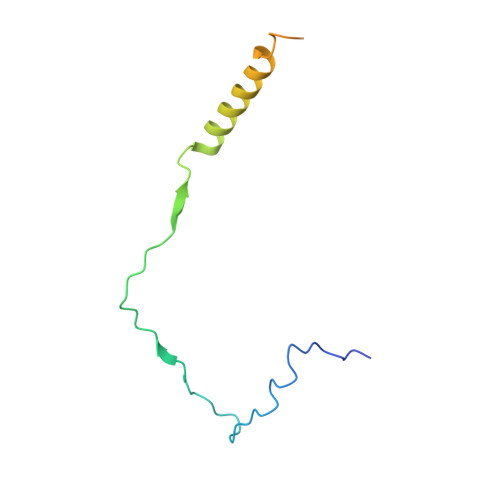
Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex.
Bussiere, D.E., Xie, L., Srinivas, H., Shu, W., Burke, A., Be, C., Zhao, J., Godbole, A., King, D., Karki, R.G., Hornak, V., Xu, F., Cobb, J., Carte, N., Frank, A.O., Frommlet, A., Graff, P., Knapp, M., Fazal, A., Okram, B., Jiang, S., Michellys, P.Y., Beckwith, R., Voshol, H., Wiesmann, C., Solomon, J.M., Paulk, J.(2020) Nat Chem Biol 16: 15-23
- PubMed: 31819272
- DOI: https://doi.org/10.1038/s41589-019-0411-6
- Primary Citation of Related Structures:
6SJ7, 6UD7, 6UE5 - PubMed Abstract:
The anticancer agent indisulam inhibits cell proliferation by causing degradation of RBM39, an essential mRNA splicing factor. Indisulam promotes an interaction between RBM39 and the DCAF15 E3 ligase substrate receptor, leading to RBM39 ubiquitination and proteasome-mediated degradation. To delineate the precise mechanism by which indisulam mediates the DCAF15-RBM39 interaction, we solved the DCAF15-DDB1-DDA1-indisulam-RBM39(RRM2) complex structure to a resolution of 2.3 Å. DCAF15 has a distinct topology that embraces the RBM39(RRM2) domain largely via non-polar interactions, and indisulam binds between DCAF15 and RBM39(RRM2), coordinating additional interactions between the two proteins. Studies with RBM39 point mutants and indisulam analogs validated the structural model and defined the RBM39 α-helical degron motif. The degron is found only in RBM23 and RBM39, and only these proteins were detectably downregulated in indisulam-treated HCT116 cells. This work further explains how indisulam induces RBM39 degradation and defines the challenge of harnessing DCAF15 to degrade additional targets.
Organizational Affiliation:
Novartis Institutes for Biomedical Research, Emeryville, CA, USA. dirksen.bussiere@novartis.com.