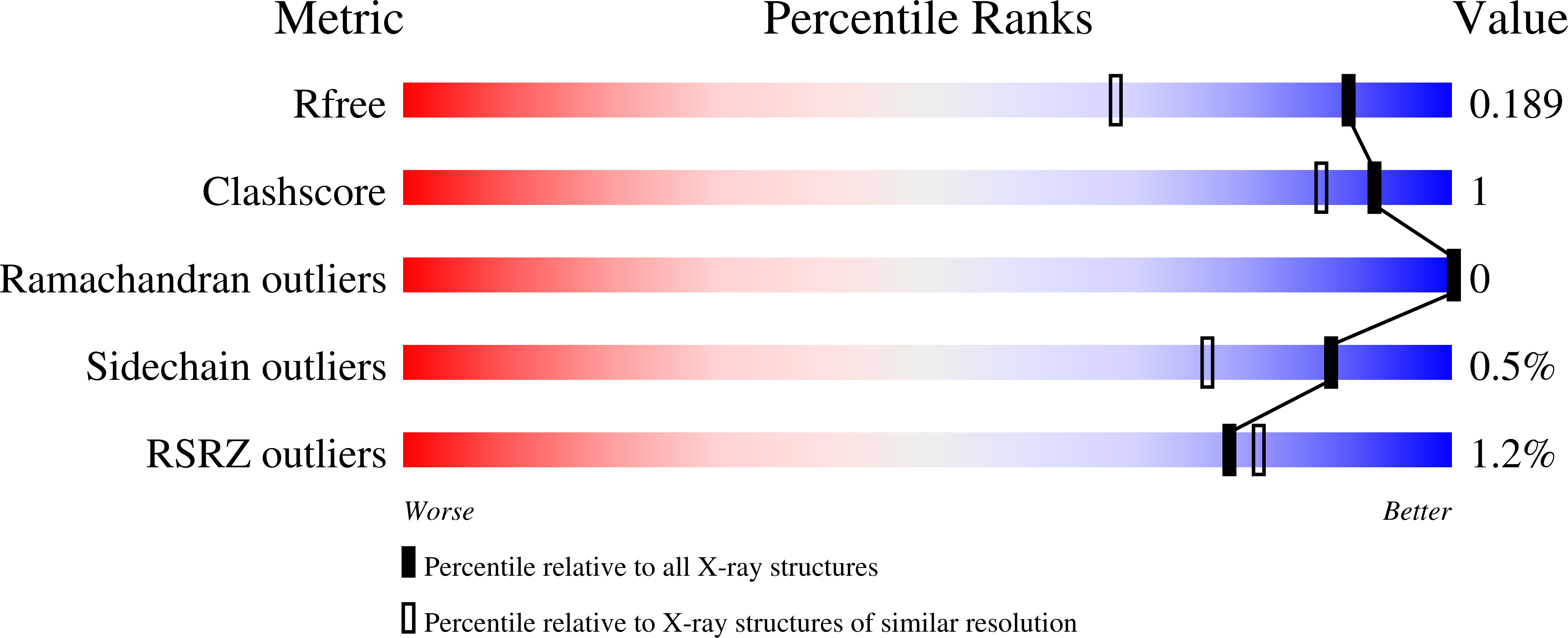
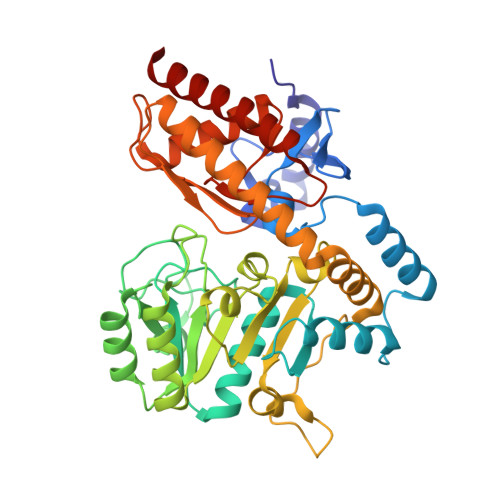
PMP-diketopiperazine adducts form at the active site of a PLP dependent enzyme involved in formycin biosynthesis.
Gao, S., Liu, H., de Crecy-Lagard, V., Zhu, W., Richards, N.G.J., Naismith, J.H.(2019) Chem Commun (Camb) 55: 14502-14505
- PubMed: 31730149
- DOI: https://doi.org/10.1039/c9cc06975e
- Primary Citation of Related Structures:
6SSD, 6SSE, 6SSF, 6SSG - PubMed Abstract:
ForI is a PLP-dependent enzyme from the biosynthetic pathway of the C-nucleoside antibiotic formycin. Cycloserine is thought to inhibit PLP-dependent enzymes by irreversibly forming a PMP-isoxazole. We now report that ForI forms novel PMP-diketopiperazine derivatives following incubation with both d and l cycloserine. This unexpected result suggests chemical diversity in the chemistry of cycloserine inhibition.
Organizational Affiliation:
Research Complex at Harwell, Didcot, OX11 0FA, UK.