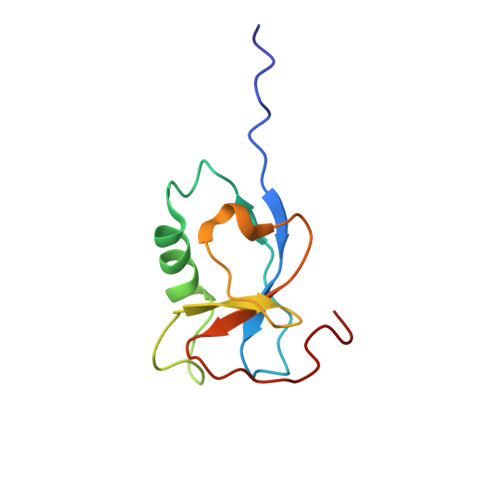
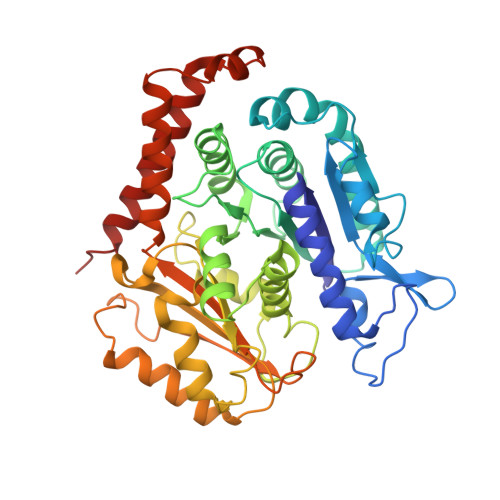
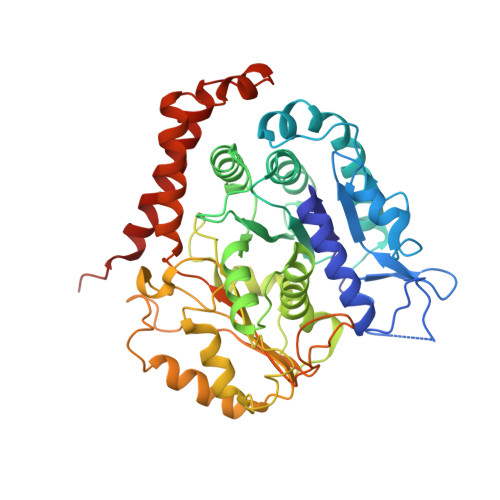
A microtubule RELION-based pipeline for cryo-EM image processing.
Cook, A.D., Manka, S.W., Wang, S., Moores, C.A., Atherton, J.(2020) J Struct Biol 209: 107402-107402
- PubMed: 31610239
- DOI: https://doi.org/10.1016/j.jsb.2019.10.004
- Primary Citation of Related Structures:
6RF8 - PubMed Abstract:
Microtubules are polar filaments built from αβ-tubulin heterodimers that exhibit a range of architectures in vitro and in vivo. Tubulin heterodimers are arranged helically in the microtubule wall but many physiologically relevant architectures exhibit a break in helical symmetry known as the seam. Noisy 2D cryo-electron microscopy projection images of pseudo-helical microtubules therefore depict distinct but highly similar views owing to the high structural similarity of α- and β-tubulin. The determination of the αβ-tubulin register and seam location during image processing is essential for alignment accuracy that enables determination of biologically relevant structures. Here we present a pipeline designed for image processing and high-resolution reconstruction of cryo-electron microscopy microtubule datasets, based in the popular and user-friendly RELION image-processing package, Microtubule RELION-based Pipeline (MiRP). The pipeline uses a combination of supervised classification and prior knowledge about geometric lattice constraints in microtubules to accurately determine microtubule architecture and seam location. The presented method is fast and semi-automated, producing near-atomic resolution reconstructions with test datasets that contain a range of microtubule architectures and binding proteins.
Organizational Affiliation:
Institute of Structural and Molecular Biology, Birkbeck, University of London, Malet Street, London, United Kingdom.