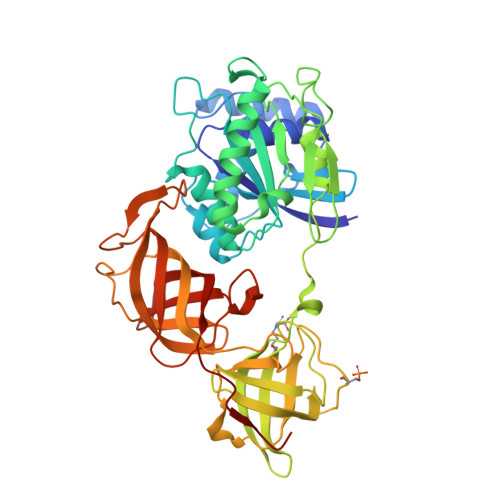
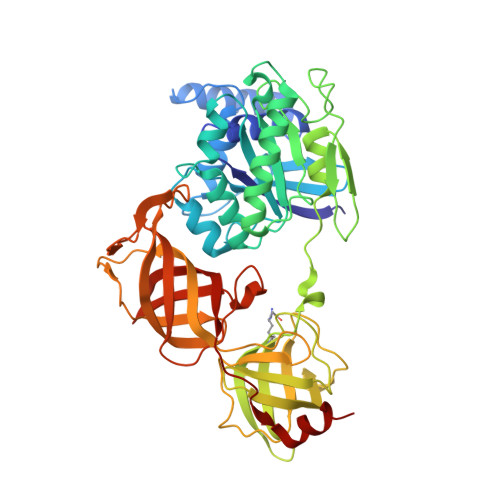
Structural Cues for Understanding eEF1A2 Moonlighting.
Carriles, A.A., Mills, A., Munoz-Alonso, M.J., Gutierrez, D., Dominguez, J.M., Hermoso, J.A., Gago, F.(2021) Chembiochem 22: 374-391
- PubMed: 32875694
- DOI: https://doi.org/10.1002/cbic.202000516
- Primary Citation of Related Structures:
6RA9 - PubMed Abstract:
Spontaneous mutations in the EEF1A2 gene cause epilepsy and severe neurological disabilities in children. The crystal structure of eEF1A2 protein purified from rabbit skeletal muscle reveals a post-translationally modified dimer that provides information about the sites of interaction with numerous binding partners, including itself, and maps these mutations onto the dimer and tetramer interfaces. The spatial locations of the side chain carboxylates of Glu301 and Glu374, to which phosphatidylethanolamine is uniquely attached via an amide bond, define the anchoring points of eEF1A2 to cellular membranes and interorganellar membrane contact sites. Additional bioinformatic and molecular modeling results provide novel structural insight into the demonstrated binding of eEF1A2 to SH3 domains, the common MAPK docking groove, filamentous actin, and phosphatidylinositol-4 kinase IIIβ. In this new light, the role of eEF1A2 as an ancient, multifaceted, and articulated G protein at the crossroads of autophagy, oncogenesis and viral replication appears very distant from the "canonical" one of delivering aminoacyl-tRNAs to the ribosome that has dominated the scene and much of the thinking for many decades.
Organizational Affiliation:
Department of Crystallography and Structural Biology, Institute of Physical-Chemistry "Rocasolano" CSIC, 28006, Madrid, Spain.