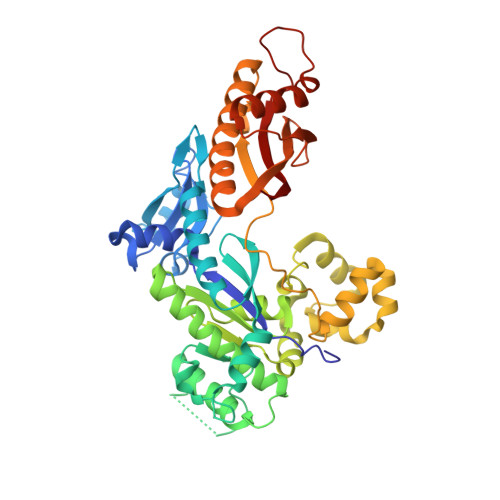
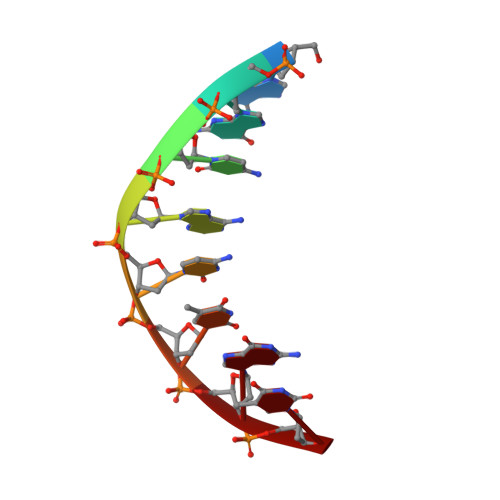
Structural insights into mutagenicity of anticancer nucleoside analog cytarabine during replication by DNA polymerase eta.
Rechkoblit, O., Johnson, R.E., Buku, A., Prakash, L., Prakash, S., Aggarwal, A.K.(2019) Sci Rep 9: 16400-16400
- PubMed: 31704958
- DOI: https://doi.org/10.1038/s41598-019-52703-7
- Primary Citation of Related Structures:
6PZ3, 6Q02 - PubMed Abstract:
Cytarabine (AraC) is the mainstay chemotherapy for acute myeloid leukemia (AML). Whereas initial treatment with AraC is usually successful, most AML patients tend to relapse, and AraC treatment-induced mutagenesis may contribute to the development of chemo-resistant leukemic clones. We show here that whereas the high-fidelity replicative polymerase Polδ is blocked in the replication of AraC, the lower-fidelity translesion DNA synthesis (TLS) polymerase Polη is proficient, inserting both correct and incorrect nucleotides opposite a template AraC base. Furthermore, we present high-resolution crystal structures of human Polη with a template AraC residue positioned opposite correct (G) and incorrect (A) incoming deoxynucleotides. We show that Polη can accommodate local perturbation caused by the AraC via specific hydrogen bonding and maintain a reaction-ready active site alignment for insertion of both correct and incorrect incoming nucleotides. Taken together, the structures provide a novel basis for the ability of Polη to promote AraC induced mutagenesis in relapsed AML patients.
Organizational Affiliation:
Department of Pharmacological Sciences, Icahn School of Medicine at Mount Sinai, Box 1677, 1425 Madison Avenue, New York, NY, 10029, USA.