Yeast- and antibody-based tools for studying tryptophan C-mannosylation.
John, A., Jarva, M.A., Shah, S., Mao, R., Chappaz, S., Birkinshaw, R.W., Czabotar, P.E., Lo, A.W., Scott, N.E., Goddard-Borger, E.D.(2021) Nat Chem Biol 17: 428-437
- PubMed: 33542533
- DOI: https://doi.org/10.1038/s41589-020-00727-w
- Primary Citation of Related Structures:
6PLH - PubMed Abstract:
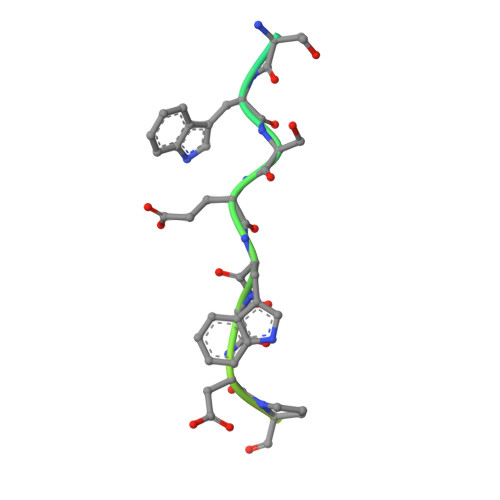
Tryptophan C-mannosylation is an unusual co-translational protein modification performed by metazoans and apicomplexan protists. The prevalence and biological functions of this modification are poorly understood, with progress in the field hampered by a dearth of convenient tools for installing and detecting the modification. Here, we engineer a yeast system to produce a diverse array of proteins with and without tryptophan C-mannosylation and interrogate the modification's influence on protein stability and function. This system also enabled mutagenesis studies to identify residues of the glycosyltransferase and its protein substrates that are crucial for catalysis. The collection of modified proteins accrued during this work facilitated the generation and thorough characterization of monoclonal antibodies against tryptophan C-mannosylation. These antibodies empowered proteomic analyses of the brain C-glycome by enriching for peptides possessing tryptophan C-mannosylation. This study revealed many new modification sites on proteins throughout the secretory pathway with both conventional and non-canonical consensus sequences.
Organizational Affiliation:
The Walter and Eliza Hall Institute of Medical Research, Parkville, Victoria, Australia.