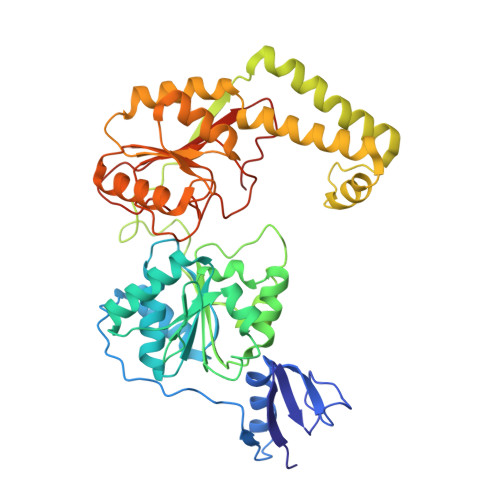
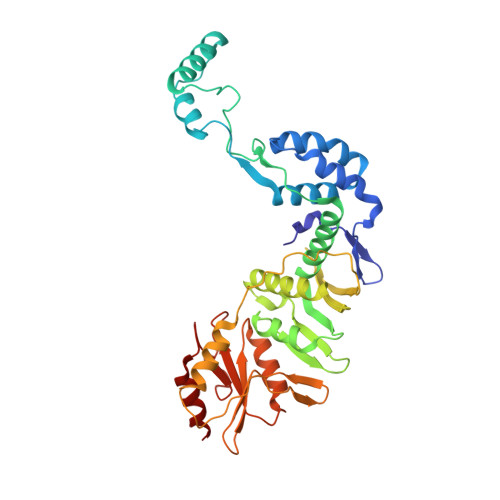
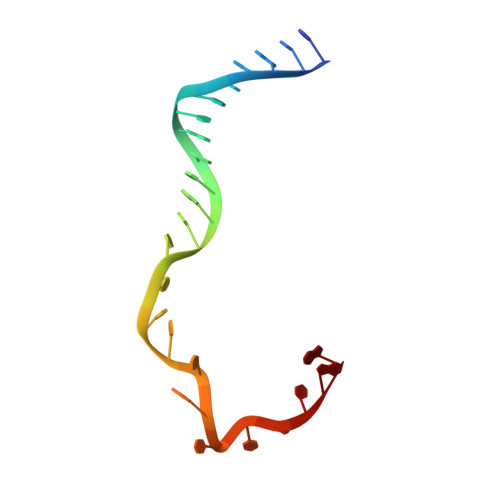
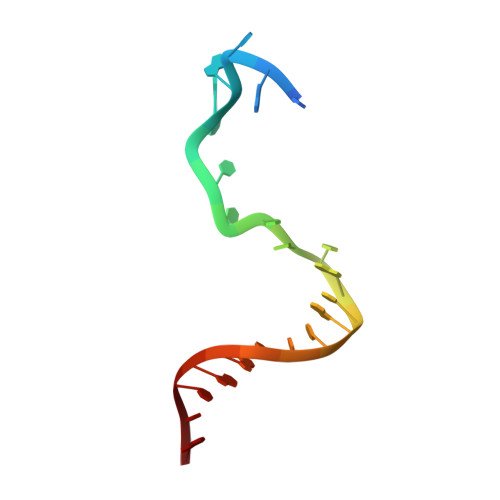
Structural basis of the XPB helicase-Bax1 nuclease complex interacting with the repair bubble DNA.
He, F., DuPrez, K., Hilario, E., Chen, Z., Fan, L.(2020) Nucleic Acids Res 48: 11695-11705
- PubMed: 32986831
- DOI: https://doi.org/10.1093/nar/gkaa801
- Primary Citation of Related Structures:
6P4F - PubMed Abstract:
Nucleotide excision repair (NER) removes various DNA lesions caused by UV light and chemical carcinogens. The DNA helicase XPB plays a key role in DNA opening and coordinating damage incision by nucleases during NER, but the underlying mechanisms remain unclear. Here, we report crystal structures of XPB from Sulfurisphaera tokodaii (St) bound to the nuclease Bax1 and their complex with a bubble DNA having one arm unwound in the crystal. StXPB and Bax1 together spirally encircle 10 base pairs of duplex DNA at the double-/single-stranded (ds-ss) junction. Furthermore, StXPB has its ThM motif intruding between the two DNA strands and gripping the 3'-overhang while Bax1 interacts with the 5'-overhang. This ternary complex likely reflects the state of repair bubble extension by the XPB and nuclease machine. ATP binding and hydrolysis by StXPB could lead to a spiral translocation along dsDNA and DNA strand separation by the ThM motif, revealing an unconventional DNA unwinding mechanism. Interestingly, the DNA is kept away from the nuclease domain of Bax1, potentially preventing DNA incision by Bax1 during repair bubble extension.
Organizational Affiliation:
Department of Biochemistry, University of California, Riverside, CA 92521, USA.