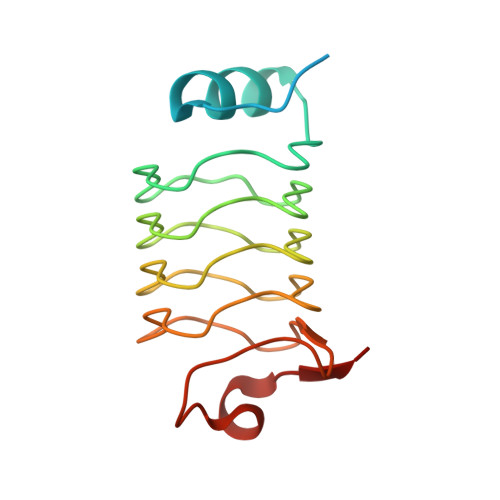
Crystal structure of Alr1298, a pentapeptide repeat protein from the cyanobacterium Nostoc sp. PCC 7120, determined at 2.1 angstrom resolution.
Zhang, R., Ni, S., Kennedy, M.A.(2020) Proteins 88: 1143-1153
- PubMed: 32092202
- DOI: https://doi.org/10.1002/prot.25882
- Primary Citation of Related Structures:
6OMX, 6UV7, 6UVI - PubMed Abstract:
Nostoc sp. PCC 7120 are filamentous cyanobacteria capable of both oxygenic photosynthesis and nitrogen fixation, with the latter taking place in specialized cells known as heterocysts that terminally differentiate from vegetative cells under conditions of nitrogen starvation. Cyanobacteria have existed on earth for more than 2 billion years and are thought to be responsible for oxygenation of the earth's atmosphere. Filamentous cyanobacteria such as Nostoc sp. PCC 7120 may also represent the oldest multicellular organisms on earth that undergo cell differentiation. Pentapeptide repeat proteins (PRPs), which occur most abundantly in cyanobacteria, adopt a right-handed quadrilateral β-helical structure, also referred to as a repeat five residue (Rfr) fold, with four-consecutive pentapeptide repeats constituting a single coil in the β-helical structure. PRPs are predicted to exist in all compartments within cyanobacteria including the thylakoid and cell-wall membranes as well as the cytoplasm and thylakoid periplasmic space. Despite their intriguing structure and importance to understanding ancient cyanobacteria, the biochemical function of PRPs in cyanobacteria remains largely unknown. Here we report the crystal structure of Alr1298, a PRP from Nostoc sp. PCC 7120 predicted to reside in the cytoplasm. The structure displays the typical right-handed quadrilateral β-helical structure and includes a four-α-helix cluster capping the N-terminus and a single α-helix capping the C-terminus. A gene cluster analysis indicated that Alr1298 may belong to an operon linked to cell proliferation and/or thylakoid biogenesis. Elevated alr1298 gene expression following nitrogen starvation indicates that Alr1298 may play a role in response to nitrogen starvation and/or heterocyst differentiation.
Organizational Affiliation:
Department of Chemistry and Biochemistry, Miami University, Oxford, Ohio.