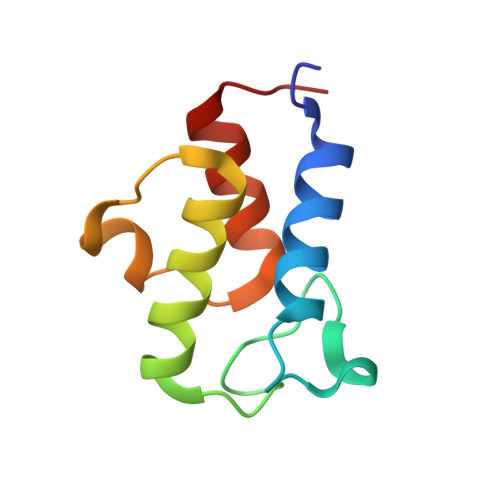
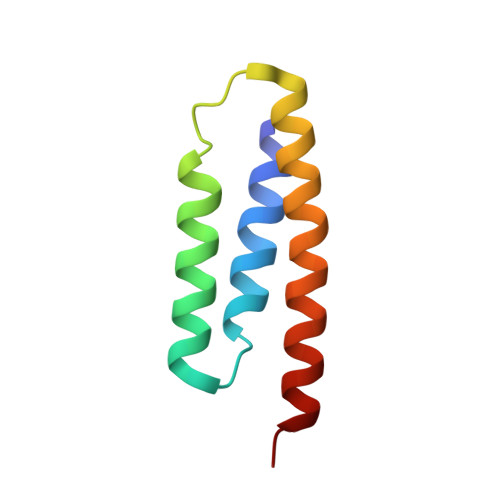
Structure of the Human ACP-ISD11 Heterodimer.
Herrera, M.G., Noguera, M.E., Sewell, K.E., Agudelo Suarez, W.A., Capece, L., Klinke, S., Santos, J.(2019) Biochemistry 58: 4596-4609
- PubMed: 31664822
- DOI: https://doi.org/10.1021/acs.biochem.9b00539
- Primary Citation of Related Structures:
6ODD - PubMed Abstract:
In recent years, the mammalian mitochondrial protein complex for iron-sulfur cluster assembly has been the focus of important studies. This is partly because of its high degree of relevance in cell metabolism and because mutations of the involved proteins are the cause of several human diseases. Cysteine desulfurase NFS1 is the key enzyme of the complex. At present, it is well-known that the active form of NFS1 is stabilized by the small protein ISD11. In this work, the structure of the human mitochondrial ACP-ISD11 heterodimer was determined at 2.0 Å resolution. ACP-ISD11 forms a cooperative unit stabilized by several ionic interactions, hydrogen bonds, and apolar interactions. The 4'-phosphopantetheine-acyl chain, which is covalently bound to ACP, interacts with several residues of ISD11, modulating together with ACP the foldability of ISD11. Recombinant human ACP-ISD11 was able to interact with the NFS1 desulfurase, thus yielding an active enzyme, and the NFS1/ACP-ISD11 core complex was activated by frataxin and ISCU proteins. Internal motions of ACP-ISD11 were studied by molecular dynamics simulations, showing the persistence of the interactions between both protein chains. The conformation of the dimer is similar to that found in the context of the (NFS1/ACP-ISD11) 2 supercomplex core, which contains the Escherichia coli ACP instead of the human variant. This fact suggests a sequential mechanism for supercomplex consolidation, in which the ACP-ISD11 complex may fold independently and, after that, the NFS1 dimer would be stabilized.
Organizational Affiliation:
Departamento de Fisiología y Biología Molecular y Celular, Facultad de Ciencias Exactas y Naturales , Universidad de Buenos Aires, Instituto de Biociencias, Biotecnología y Biomedicina (iB3), Intendente Güiraldes 2160-Ciudad Universitaria , C1428EGA Buenos Aires , Argentina.