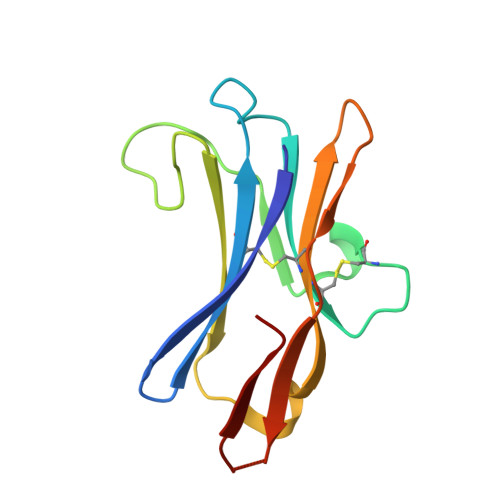
Structural Basis of CD160:HVEM Recognition.
Liu, W., Garrett, S.C., Fedorov, E.V., Ramagopal, U.A., Garforth, S.J., Bonanno, J.B., Almo, S.C.(2019) Structure 27: 1286-1295.e4
- PubMed: 31230945
- DOI: https://doi.org/10.1016/j.str.2019.05.010
- Primary Citation of Related Structures:
6NG9, 6NGG - PubMed Abstract:
CD160 is a signaling molecule that interacts with herpes virus entry mediator (HVEM) and contributes to a wide range of immune responses, including T cell inhibition, natural killer cell activation, and mucosal immunity. GPI-anchored and transmembrane isoforms of CD160 share the same ectodomain responsible for HVEM engagement, which leads to bidirectional signaling. Despite the importance of the CD160:HVEM signaling axis and its therapeutic relevance, the structural and mechanistic basis underlying CD160-HVEM engagement has not been described. We report the crystal structures of the human CD160 extracellular domain and its complex with human HVEM. CD160 adopts a unique variation of the immunoglobulin fold and exists as a monomer in solution. The CD160:HVEM assembly exhibits a 1:1 stoichiometry and a binding interface similar to that observed in the BTLA:HVEM complex. Our work reveals the chemical and physical determinants underlying CD160:HVEM recognition and initiation of associated signaling processes.
Organizational Affiliation:
Department of Biochemistry, Albert Einstein College of Medicine, 1300 Morris Park Avenue, Bronx, NY 10461, USA.