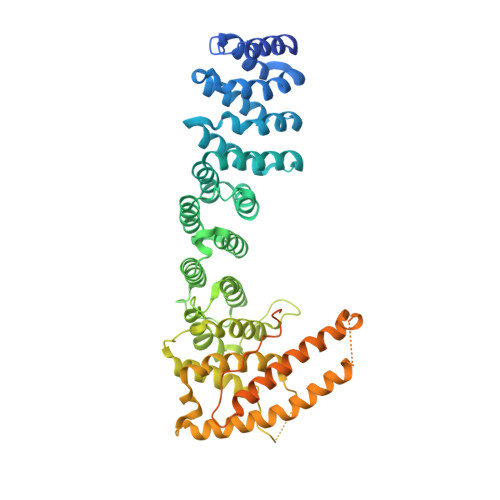
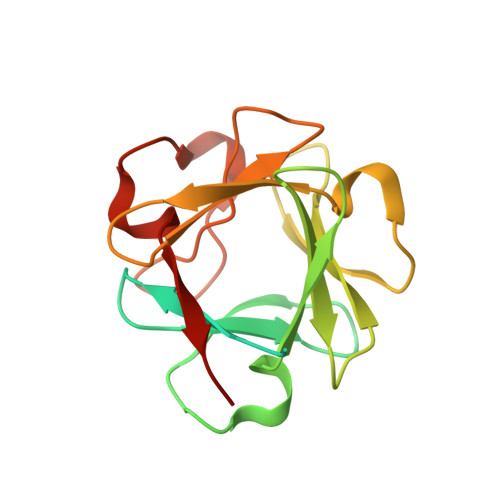
Regulation of mRNA export through API5 and nuclear FGF2 interaction.
Bong, S.M., Bae, S.H., Song, B., Gwak, H., Yang, S.W., Kim, S., Nam, S., Rajalingam, K., Oh, S.J., Kim, T.W., Park, S., Jang, H., Lee, B.I.(2020) Nucleic Acids Res 48: 6340-6352
- PubMed: 32383752
- DOI: https://doi.org/10.1093/nar/gkaa335
- Primary Citation of Related Structures:
6L4O - PubMed Abstract:
API5 (APoptosis Inhibitor 5) and nuclear FGF2 (Fibroblast Growth Factor 2) are upregulated in various human cancers and are correlated with poor prognosis. Although their physical interaction has been identified, the function related to the resulting complex is unknown. Here, we determined the crystal structure of the API5-FGF2 complex and identified critical residues driving the protein interaction. These findings provided a structural basis for the nuclear localization of the FGF2 isoform lacking a canonical nuclear localization signal and identified a cryptic nuclear localization sequence in FGF2. The interaction between API5 and FGF2 was important for mRNA nuclear export through both the TREX and eIF4E/LRPPRC mRNA export complexes, thus regulating the export of bulk mRNA and specific mRNAs containing eIF4E sensitivity elements, such as c-MYC and cyclin D1. These data show the newly identified molecular function of API5 and nuclear FGF2, and provide a clue to understanding the dynamic regulation of mRNA export.
Organizational Affiliation:
Research Institute, National Cancer Center, Goyang-si, Gyeonggi 10408, Republic of Korea.