The crystal structure of light-driven cyanobacterial chloride importer from Mastigocladopsis repens
Yun, J.H., Park, J.H., Jin, Z., Ohki, M., Wang, Y., Lupala, C.S., Kim, M., Liu, H., Park, S.Y., Lee, W.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cyanobacterial chloride importer | 232 | Mastigocladopsis repens | Mutation(s): 0 | 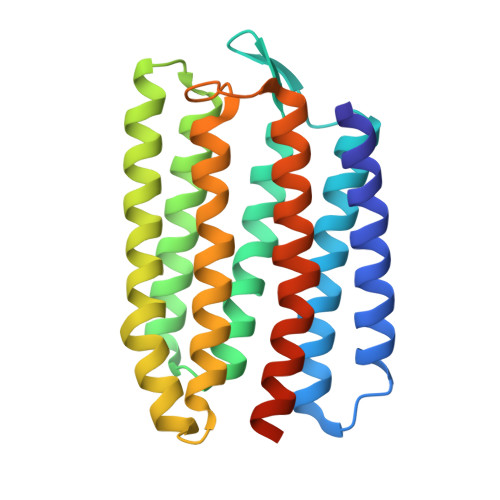 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| RET Query on RET | B [auth A] | RETINAL C20 H28 O NCYCYZXNIZJOKI-OVSJKPMPSA-N |  | ||
| OLA (Subject of Investigation/LOI) Query on OLA | C [auth A] D [auth A] E [auth A] F [auth A] G [auth A] | OLEIC ACID C18 H34 O2 ZQPPMHVWECSIRJ-KTKRTIGZSA-N |  | ||
| CL Query on CL | Q [auth A], R [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 61.685 | α = 90 |
| b = 61.685 | β = 90 |
| c = 240.466 | γ = 120 |
| Software Name | Purpose |
|---|---|
| BUSTER | refinement |
| MxDC | data collection |
| HKL-2000 | data scaling |
| PHENIX | phasing |
| Coot | model building |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Research Foundation (Korea) | Korea, Republic Of | 20171A2B2008483 |
| National Research Foundation (Korea) | Korea, Republic Of | 20161A6A3A04010213 |