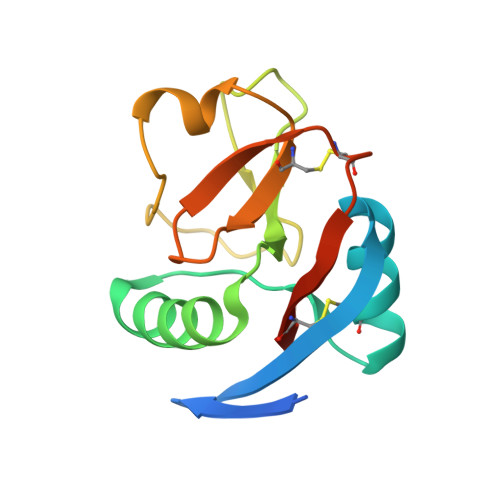
Crystal structure of the CTLD7 domain of human M-type phospholipase A2 receptor.
Yu, B., Hu, Z., Kong, D., Cheng, C., He, Y.(2019) J Struct Biol 207: 295-300
- PubMed: 31271865
- DOI: https://doi.org/10.1016/j.jsb.2019.06.007
- Primary Citation of Related Structures:
6JLI - PubMed Abstract:
M-type phospholipase A2 receptor (PLA2R) is a member of the mannose receptor family. Recent evidence shows that PLA2R is a major autoantigen causing idiopathic membranous nephropathy (IMN), which is an autoimmune disease and one of the most common causes for nephrotic syndrome in adults. The epitope mapping data suggest that the major epitopes of PLA2R locate at the CysR, CTLD1 and CTLD7 domains. However, due to the lack of the high-resolution structural information, it is unclear how the autoantibodies interact with PLA2R. Here we determine the crystal structure of the CTLD7 domain of PLA2R at 1.8 Å, showing that it adopts a typical CTLD fold, and the structural alignments also provide hints for the potential antibody binding regions. In addition, the high-resolution structural information of CTLD7 could be applied to identify the epitopes for autoantibodies, which would facilitate the therapeutic strategies against IMN.
Organizational Affiliation:
National Center for Protein Science Shanghai, Shanghai Science Research Center; CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences; University of Chinese Academy of Sciences, Shanghai, China.