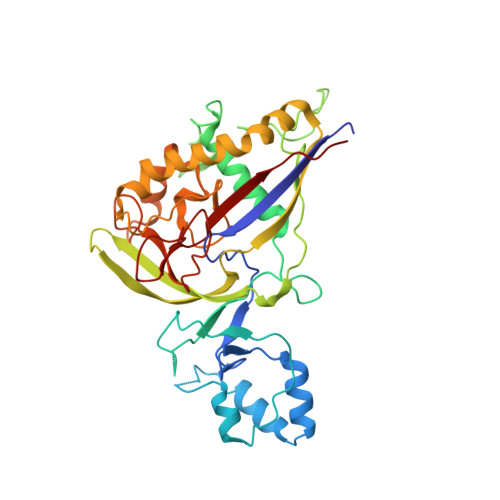
Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
You, L., Ma, J., Wang, J., Artamonova, D., Wang, M., Liu, L., Xiang, H., Severinov, K., Zhang, X., Wang, Y.(2019) Cell 176: 239-253.e16
- PubMed: 30503210
- DOI: https://doi.org/10.1016/j.cell.2018.10.052
- Primary Citation of Related Structures:
6IFK, 6IFL, 6IFN, 6IFR, 6IFU, 6IFY, 6IFZ, 6IG0 - PubMed Abstract:
Csm, a type III-A CRISPR-Cas interference complex, is a CRISPR RNA (crRNA)-guided RNase that also possesses target RNA-dependent DNase and cyclic oligoadenylate (cOA) synthetase activities. However, the structural features allowing target RNA-binding-dependent activation of DNA cleavage and cOA generation remain unknown. Here, we report the structure of Csm in complex with crRNA together with structures of cognate or non-cognate target RNA bound Csm complexes. We show that depending on complementarity with the 5' tag of crRNA, the 3' anti-tag region of target RNA binds at two distinct sites of the Csm complex. Importantly, the interaction between the non-complementary anti-tag region of cognate target RNA and Csm1 induces a conformational change at the Csm1 subunit that allosterically activates DNA cleavage and cOA generation. Together, our structural studies provide crucial insights into the mechanistic processes required for crRNA-meditated sequence-specific RNA cleavage, RNA target-dependent non-specific DNA cleavage, and cOA generation.
Organizational Affiliation:
Key Laboratory of RNA Biology, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, 100101 Beijing, China; University of Chinese Academy of Sciences, 100049 Beijing, China.