Crystal structure of the R460G disease-causing mutant of the human dihydrolipoamide dehydrogenase.
Ambrus, A., Szabo, E., Weichsel, A., Bui, D., Torocsik, B., Montfort, W.R., Jordan, F., Adam-Vizi, V.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Dihydrolipoyl dehydrogenase, mitochondrial | 496 | Homo sapiens | Mutation(s): 1 Gene Names: DLD, GCSL, LAD, PHE3 EC: 1.8.1.4 | 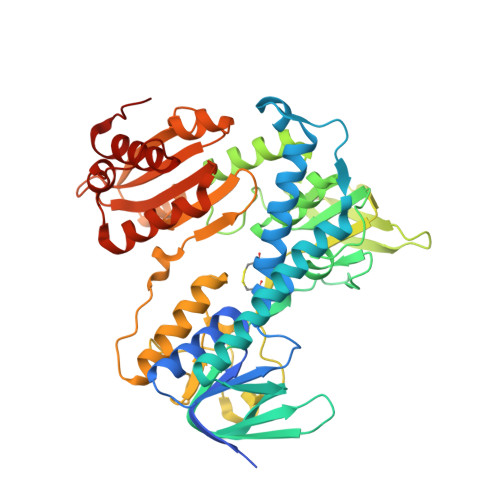 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P09622 (Homo sapiens) Explore P09622 Go to UniProtKB: P09622 | |||||
PHAROS: P09622 GTEx: ENSG00000091140 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P09622 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FAD Query on FAD | C [auth A], J [auth B] | FLAVIN-ADENINE DINUCLEOTIDE C27 H33 N9 O15 P2 VWWQXMAJTJZDQX-UYBVJOGSSA-N |  | ||
| TRS Query on TRS | I [auth A] | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL C4 H12 N O3 LENZDBCJOHFCAS-UHFFFAOYSA-O |  | ||
| SO4 Query on SO4 | D [auth A] E [auth A] F [auth A] G [auth A] H [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 117.913 | α = 90 |
| b = 169.497 | β = 90 |
| c = 61.401 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| MxCuBE | data collection |
| XDS | data scaling |
| XDS | data reduction |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Hungary | Hungarian Academy of Sciences (MTA), grant#: 02001 [to V.A.-V.] | |
| Hungary | Hungarian Scientific Research Fund (OTKA), grant#: 112230 [to V.A.-V.] | |
| Hungary | Hungarian Brain Research Program (NAP), grant#: 2017-1.2.1-NKP2017-00002 [to V.A.-V.] | |
| European Molecular Biology Organization | Germany | Short-term Fellowship [to A.A.] |
| Hungary | Hungarian Academy of Sciences, Bolyai Fellowship [to A.A] | |
| Hungary | Semmelweis University, Young Investigator Research Grant [to A.A.] | |
| Hungary | Gedeon Richter Pharmaceuticals PIc., Young Investigator Research Grant [to A.A.] | |
| Germany | European Union, Horizon 2020 Research and Innovation Programme grant [to A.A.] |