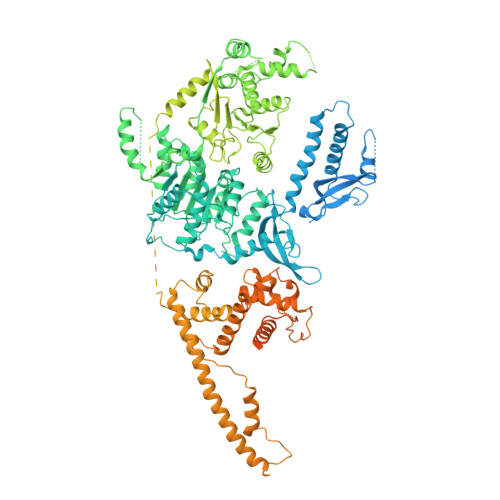
Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome.
Sundaramoorthy, R., Hughes, A.L., El-Mkami, H., Norman, D.G., Ferreira, H., Owen-Hughes, T.(2018) Elife 7
- PubMed: 30079888
- DOI: https://doi.org/10.7554/eLife.35720
- Primary Citation of Related Structures:
6FTX, 6G0L - PubMed Abstract:
ATP-dependent chromatin remodelling proteins represent a diverse family of proteins that share ATPase domains that are adapted to regulate protein-DNA interactions. Here, we present structures of the Saccharomyces cerevisiae Chd1 protein engaged with nucleosomes in the presence of the transition state mimic ADP-beryllium fluoride. The path of DNA strands through the ATPase domains indicates the presence of contacts conserved with single strand translocases and additional contacts with both strands that are unique to Snf2 related proteins. The structure provides connectivity between rearrangement of ATPase lobes to a closed, nucleotide bound state and the sensing of linker DNA. Two turns of linker DNA are prised off the surface of the histone octamer as a result of Chd1 binding, and both the histone H3 tail and ubiquitin conjugated to lysine 120 are re-orientated towards the unravelled DNA. This indicates how changes to nucleosome structure can alter the way in which histone epitopes are presented.
Organizational Affiliation:
Centre for Gene Regulation and Expression, School of Life Sciences, University of Dundee, Dundee, United Kingdom.