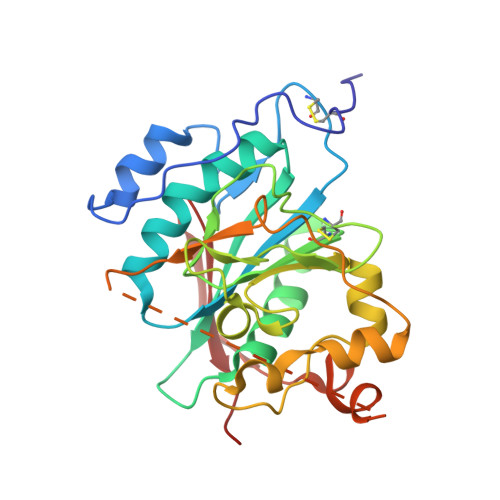
The dimeric structure of wild-type human glycosyltransferase B4GalT1.
Harrus, D., Khoder-Agha, F., Peltoniemi, M., Hassinen, A., Ruddock, L., Kellokumpu, S., Glumoff, T.(2018) PLoS One 13: e0205571-e0205571
- PubMed: 30352055
- DOI: https://doi.org/10.1371/journal.pone.0205571
- Primary Citation of Related Structures:
6FWT, 6FWU - PubMed Abstract:
Most glycosyltransferases, including B4GalT1 (EC 2.4.1.38), are known to assemble into enzyme homomers and functionally relevant heteromers in vivo. However, it remains unclear why and how these enzymes interact at the molecular/atomic level. Here, we solved the crystal structure of the wild-type human B4GalT1 homodimer. We also show that B4GalT1 exists in a dynamic equilibrium between monomer and dimer, since a purified monomer reappears as a mixture of both and as we obtained crystal forms of the monomer and dimer assemblies in the same crystallization conditions. These two crystal forms revealed the unliganded B4GalT1 in both the open and the closed conformation of the Trp loop and the lid regions, responsible for donor and acceptor substrate binding, respectively. The present structures also show the lid region in full in an open conformation, as well as a new conformation for the GlcNAc acceptor loop (residues 272-288). The physiological relevance of the homodimer in the crystal was validated by targeted mutagenesis studies coupled with FRET assays. These showed that changing key catalytic amino acids impaired homomer formation in vivo. The wild-type human B4GalT1 structure also explains why the variant proteins used for crystallization in earlier studies failed to reveal the homodimers described in this study.
Organizational Affiliation:
Faculty of Biochemistry and Molecular Medicine, University of Oulu, Aapistie 7A, Oulu, Finland.