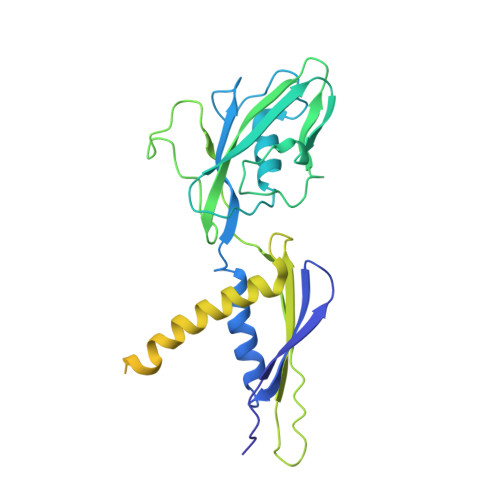
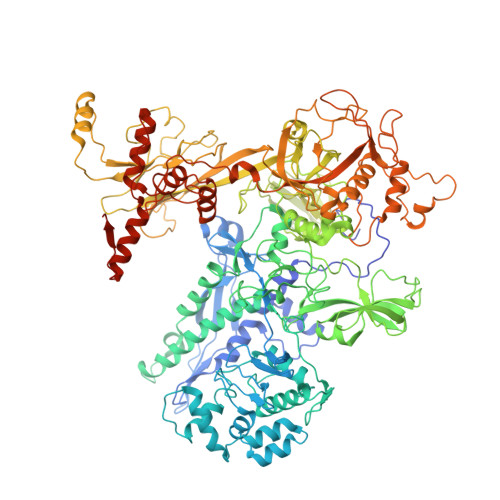
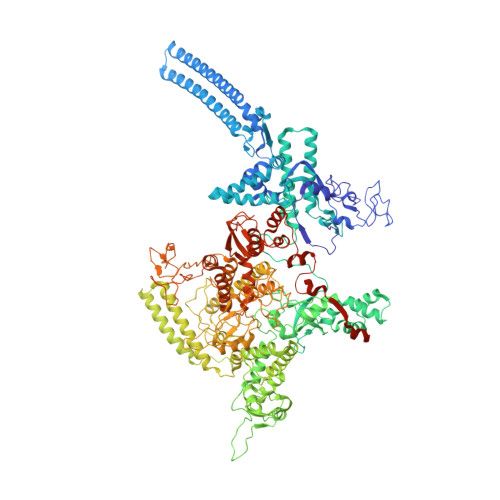
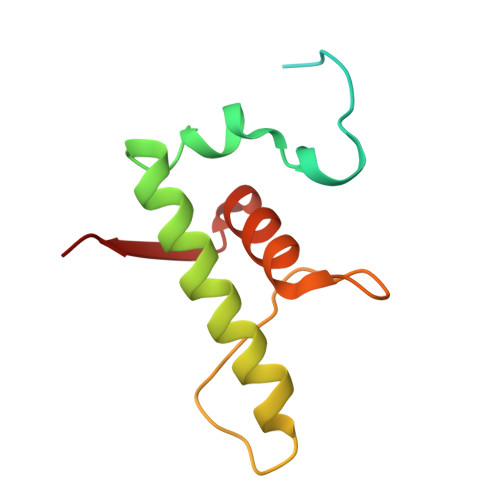
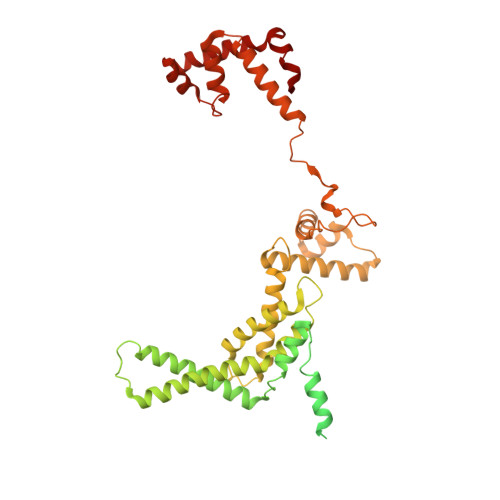
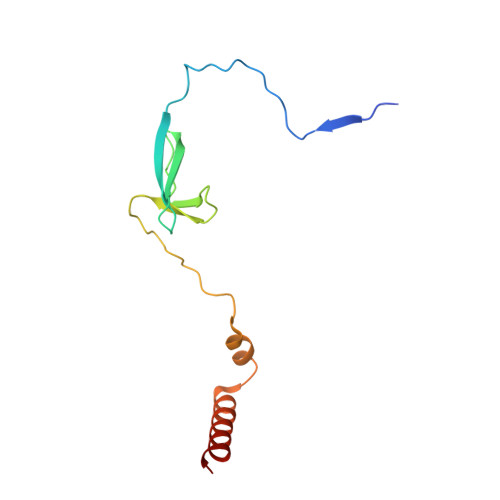
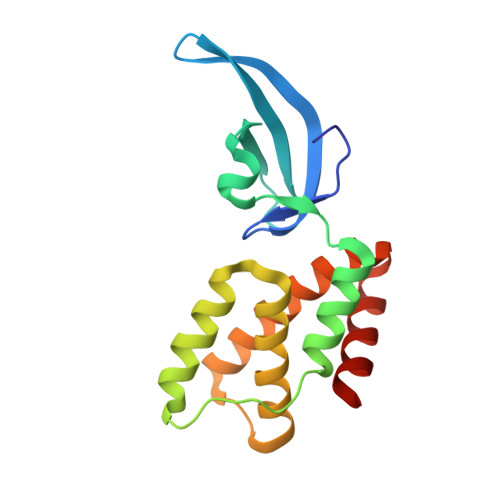
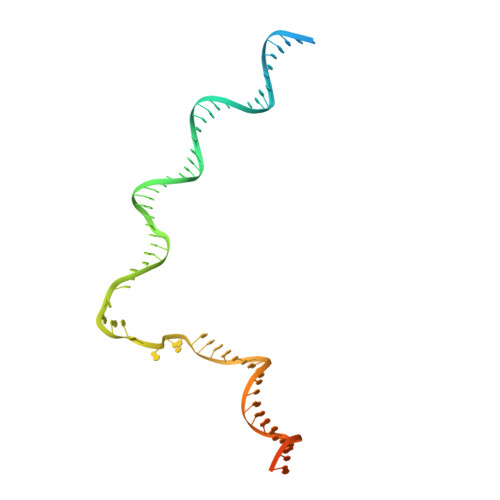
Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding.
Boyaci, H., Chen, J., Jansen, R., Darst, S.A., Campbell, E.A.(2019) Nature 565: 382-385
- PubMed: 30626968
- DOI: https://doi.org/10.1038/s41586-018-0840-5
- Primary Citation of Related Structures:
6EDT, 6EE8, 6EEC, 6M7J - PubMed Abstract:
A key regulated step of transcription is promoter melting by RNA polymerase (RNAP) to form the open promoter complex 1-3 . To generate the open complex, the conserved catalytic core of the RNAP combines with initiation factors to locate promoter DNA, unwind 12-14 base pairs of the DNA duplex and load the template-strand DNA into the RNAP active site. Formation of the open complex is a multi-step process during which transient intermediates of unknown structure are formed 4-6 . Here we present cryo-electron microscopy structures of bacterial RNAP-promoter DNA complexes, including structures of partially melted intermediates. The structures show that late steps of promoter melting occur within the RNAP cleft, delineate key roles for fork-loop 2 and switch 2-universal structural features of RNAP-in restricting access of DNA to the RNAP active site, and explain why clamp opening is required to allow entry of single-stranded template DNA into the active site. The key roles of fork-loop 2 and switch 2 suggest a common mechanism for late steps in promoter DNA opening to enable gene expression across all domains of life.
Organizational Affiliation:
Laboratory of Molecular Biophysics, The Rockefeller University, New York, NY, USA.