Crystal structures of the complex of a kallikrein inhibitor from Bauhinia bauhinioides with trypsin and modeling of kallikrein complexes.
Li, M., Srp, J., Gustchina, A., Dauter, Z., Mares, M., Wlodawer, A.(2019) Acta Crystallogr D Struct Biol 75: 56-69
- PubMed: 30644845
- DOI: https://doi.org/10.1107/S2059798318016492
- Primary Citation of Related Structures:
6DWF, 6DWH, 6DWU - PubMed Abstract:
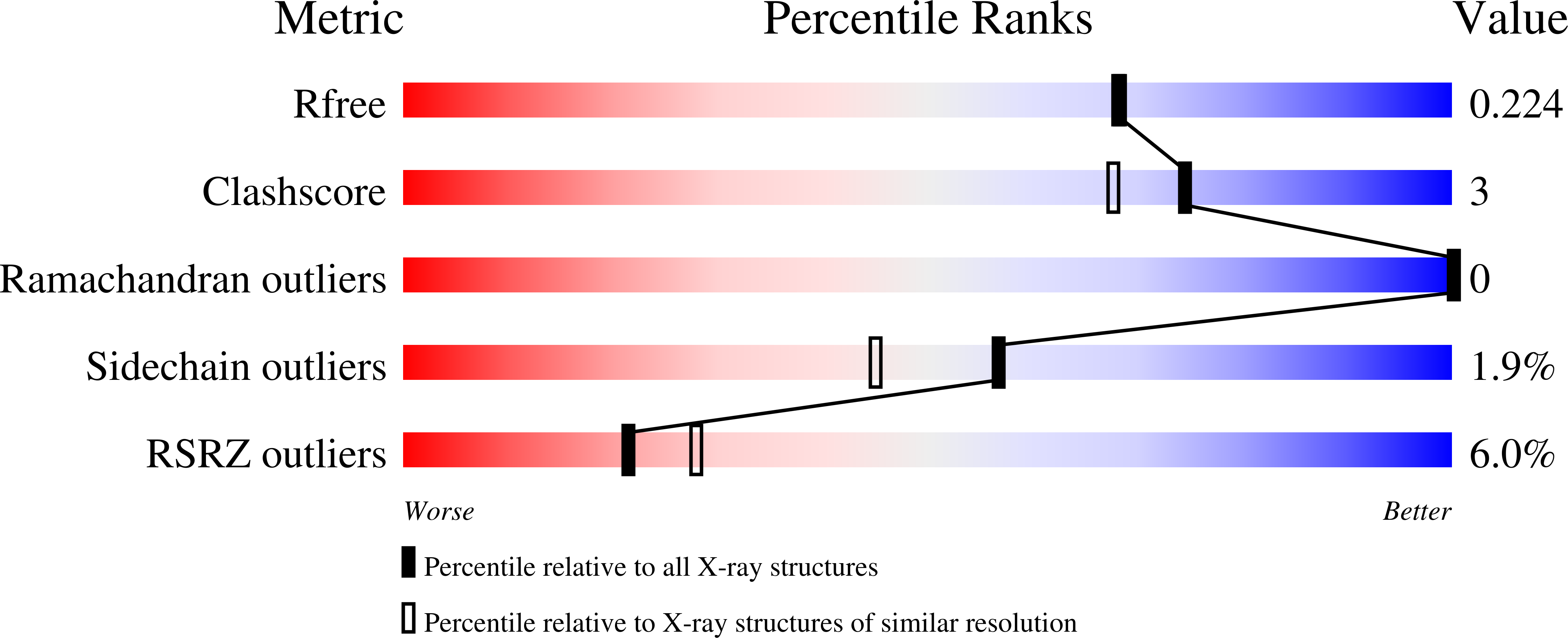
Structures of a recombinant Kunitz-type serine protease inhibitor from Bauhinia bauhinioides (BbKI) complexed with bovine trypsin were determined in two crystal forms. The crystal structure with the L55R mutant of BbKI was determined in space group P6 4 at 1.94 Å resolution and that with native BbKI in the monoclinic space group P2 1 at 3.95 Å resolution. The asymmetric unit of the latter crystals contained 44 independent complexes, thus representing one of the largest numbers of independent objects deposited in the Protein Data Bank. Additionally, the structure of the complex with native BbKI was determined at 2.0 Å resolution from P6 4 crystals isomorphous to those of the mutant. Since BbKI has previously been found to be a potent inhibitor of the trypsin-like plasma kallikrein, it was also tested against several tissue kallikreins. It was found that BbKI is a potent inhibitor of human tissue kallikrein 4 (KLK4) and the chymotrypsin-like human tissue kallikrein 7 (KLK7). Structures of BbKI complexed with the catalytic domain of human plasma kallikrein were modeled, as well as those with KLK4 and KLK7, and the structures were analyzed in order to identify the interactions that are responsible for inhibitory potency.
Organizational Affiliation:
Macromolecular Crystallography Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702, USA.