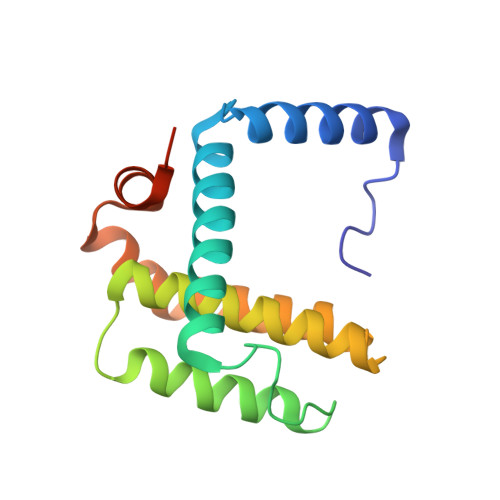
Structural insights into BCL2 pro-survival protein interactions with the key autophagy regulator BECN1 following phosphorylation by STK4/MST1.
Lee, E.F., Smith, N.A., Soares da Costa, T.P., Meftahi, N., Yao, S., Harris, T.J., Tran, S., Pettikiriarachchi, A., Perugini, M.A., Keizer, D.W., Evangelista, M., Smith, B.J., Fairlie, W.D.(2019) Autophagy 15: 785-795
- PubMed: 30626284
- DOI: https://doi.org/10.1080/15548627.2018.1564557
- Primary Citation of Related Structures:
6DCN, 6DCO - PubMed Abstract:
BECN1/Beclin 1 is a critical protein in the initiation of autophagosome formation. Recent studies have shown that phosphorylation of BECN1 by STK4/MST1 at threonine 108 (T108) within its BH3 domain blocks macroautophagy/autophagy by increasing BECN1 affinity for its negative regulators, the anti-apoptotic proteins BCL2/Bcl-2 and BCL2L1/Bcl-x L . It was proposed that this increased binding is due to formation of an electrostatic interaction with a conserved histidine residue on the anti-apoptotic molecules. Here, we performed biophysical studies which demonstrated that a peptide corresponding to the BECN1 BH3 domain in which T108 is phosphorylated (p-T108) does show increased affinity for anti-apoptotic proteins that is significant, though only minor (<2-fold). We also determined X-ray crystal structures of BCL2 and BCL2L1 with T108-modified BECN1 BH3 peptides, but only showed evidence of an interaction between the BH3 peptide and the conserved histidine residue when the histidine flexibility was restrained due to crystal contacts. These data, together with molecular dynamics studies, indicate that the histidine is highly flexible, even when complexed with BECN1 BH3. Binding studies also showed that detergent can increase the affinity of the interaction. Although this increase was similar for both the phosphorylated and non-phosphorylated peptides, it suggests factors such as membranes could impact on the interaction between BECN1 and BCL2 proteins, and therefore, on the regulation of autophagy. Hence, we propose that phosphorylation of BECN1 by STK4/MST1 can increase the affinity of the interaction between BECN1 and anti-apoptotic proteins and this interaction can be stabilized by local environmental factors. Abbreviations: asu: asymmetric unit; BH3: BCL2/Bcl-2 homology 3; DAPK: death associated protein kinase; MD: molecular dynamics; MST: microscale thermophoresis; NMR: nuclear magnetic resonance; PDB: protein data bank; p-T: phosphothreonine; SPR: surface plasmon resonance; STK4/MST1: serine/threonine kinase 4.
Organizational Affiliation:
a La Trobe Institute for Molecular Science , La Trobe University , Melbourne , Australia.