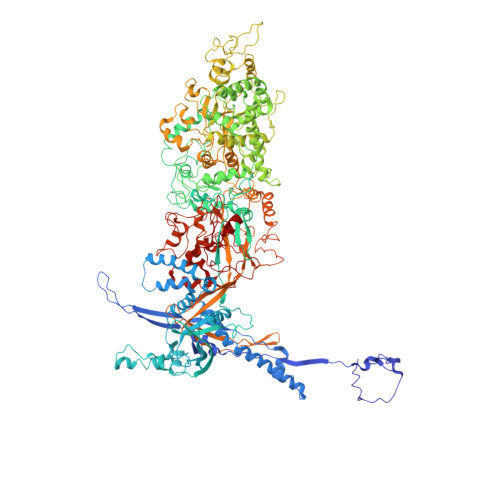
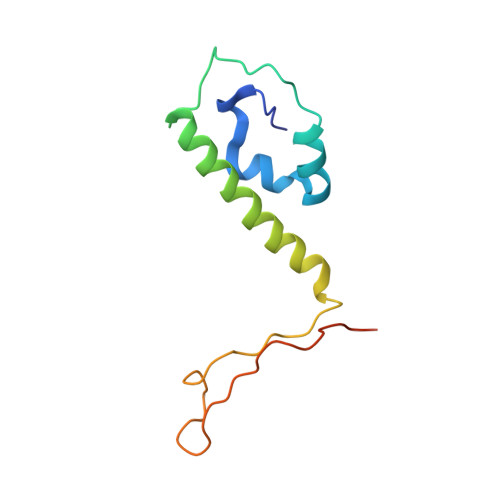
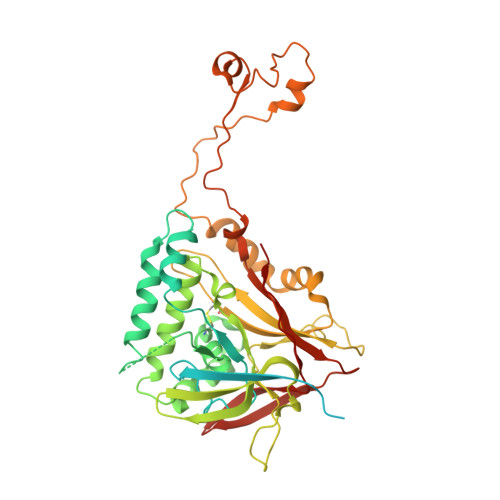
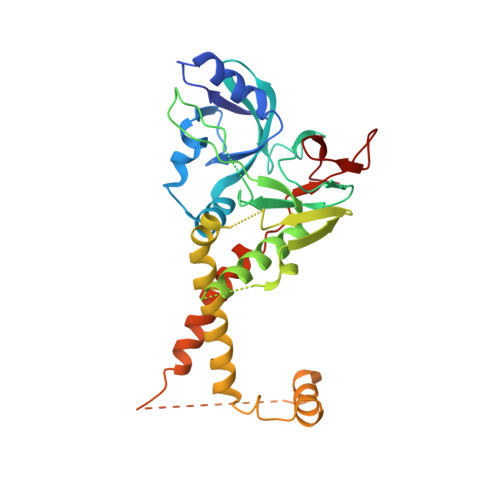
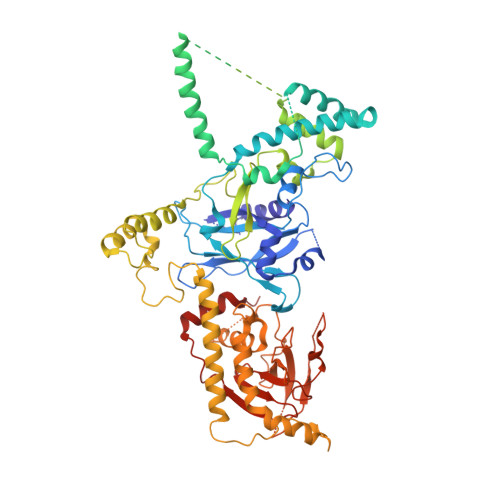
Structure of the herpes simplex virus 1 capsid with associated tegument protein complexes.
Dai, X., Zhou, Z.H.(2018) Science 360
- PubMed: 29622628
- DOI: https://doi.org/10.1126/science.aao7298
- Primary Citation of Related Structures:
6CGR - PubMed Abstract:
Herpes simplex viruses (HSVs) rely on capsid-associated tegument complex (CATC) for long-range axonal transport of their genome-containing capsids between sites of infection and neuronal cell bodies. Here we report cryo-electron microscopy structures of the HSV-1 capsid with CATC up to 3.5-angstrom resolution and atomic models of multiple conformers of capsid proteins VP5, VP19c, VP23, and VP26 and tegument proteins pUL17, pUL25, and pUL36. Crowning every capsid vertex are five copies of heteropentameric CATC, each containing a pUL17 monomer supporting the coiled-coil helix bundle of a pUL25 dimer and a pUL36 dimer, thus positioning their flexible domains for potential involvement in nuclear capsid egress and axonal capsid transport. Notwithstanding newly discovered fold conservation between triplex proteins and bacteriophage λ protein gpD and the previously recognized bacteriophage HK97 gp5-like fold in VP5, HSV-1 capsid proteins exhibit extraordinary diversity in forms of domain insertion and conformational polymorphism, not only for interactions with tegument proteins but also for encapsulation of large genomes.
Organizational Affiliation:
Department of Microbiology, Immunology, and Molecular Genetics, University of California, Los Angeles, Los Angeles, CA 90095, USA.