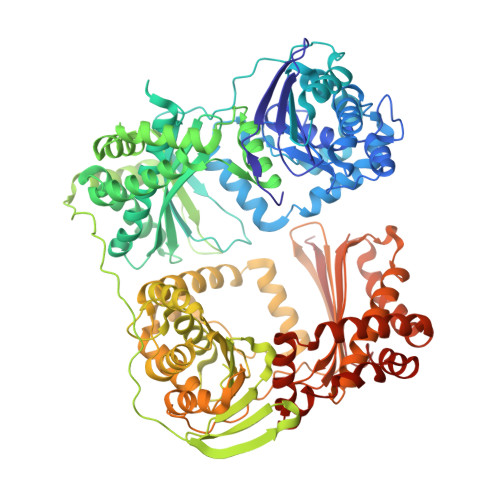
FusC, a member of the M16 protease family acquired by bacteria for iron piracy against plants.
Grinter, R., Hay, I.D., Song, J., Wang, J., Teng, D., Dhanesakaran, V., Wilksch, J.J., Davies, M.R., Littler, D., Beckham, S.A., Henderson, I.R., Strugnell, R.A., Dougan, G., Lithgow, T.(2018) PLoS Biol 16: e2006026-e2006026
- PubMed: 30071011
- DOI: https://doi.org/10.1371/journal.pbio.2006026
- Primary Citation of Related Structures:
6B03, 6B05, 6BRS - PubMed Abstract:
Iron is essential for life. Accessing iron from the environment can be a limiting factor that determines success in a given environmental niche. For bacteria, access of chelated iron from the environment is often mediated by TonB-dependent transporters (TBDTs), which are β-barrel proteins that form sophisticated channels in the outer membrane. Reports of iron-bearing proteins being used as a source of iron indicate specific protein import reactions across the bacterial outer membrane. The molecular mechanism by which a folded protein can be imported in this way had remained mysterious, as did the evolutionary process that could lead to such a protein import pathway. How does the bacterium evolve the specificity factors that would be required to select and import a protein encoded on another organism's genome? We describe here a model whereby the plant iron-bearing protein ferredoxin can be imported across the outer membrane of the plant pathogen Pectobacterium by means of a Brownian ratchet mechanism, thereby liberating iron into the bacterium to enable its growth in plant tissues. This import pathway is facilitated by FusC, a member of the same protein family as the mitochondrial processing peptidase (MPP). The Brownian ratchet depends on binding sites discovered in crystal structures of FusC that engage a linear segment of the plant protein ferredoxin. Sequence relationships suggest that the bacterial gene encoding FusC has previously unappreciated homologues in plants and that the protein import mechanism employed by the bacterium is an evolutionary echo of the protein import pathway in plant mitochondria and plastids.
Organizational Affiliation:
Infection and Immunity Program, Biomedicine Discovery Institute and Department of Microbiology, Monash University, Clayton, Australia.