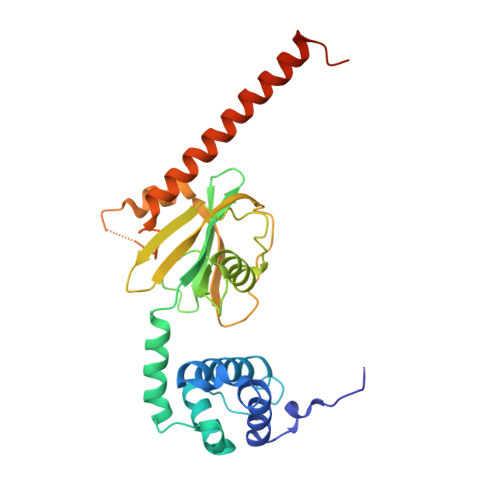
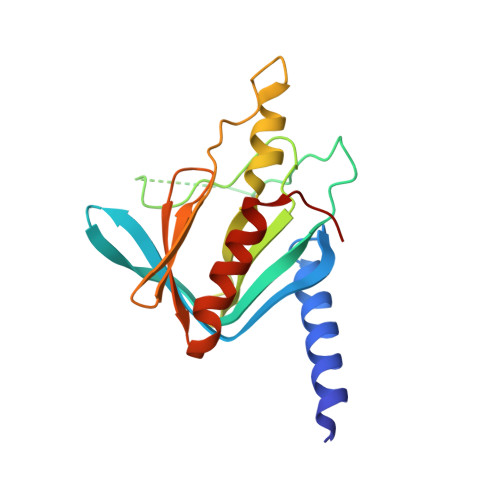
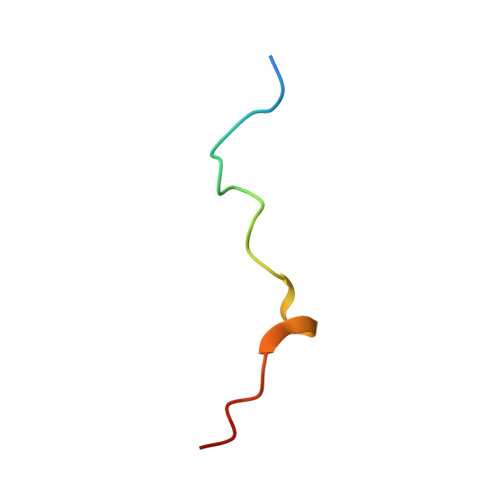
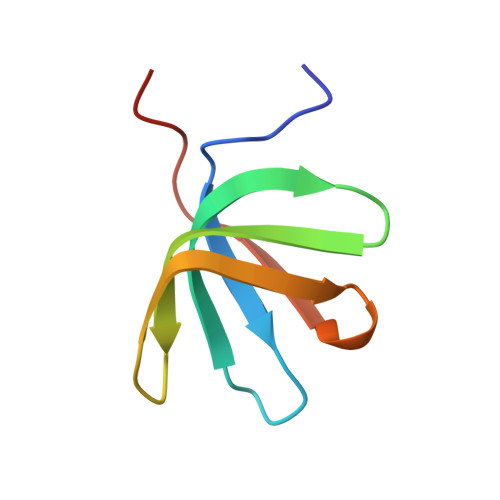
Structure of the activated Edc1-Dcp1-Dcp2-Edc3 mRNA decapping complex with substrate analog poised for catalysis.
Mugridge, J.S., Tibble, R.W., Ziemniak, M., Jemielity, J., Gross, J.D.(2018) Nat Commun 9: 1152-1152
- PubMed: 29559651
- DOI: https://doi.org/10.1038/s41467-018-03536-x
- Primary Citation of Related Structures:
6AM0 - PubMed Abstract:
The conserved decapping enzyme Dcp2 recognizes and removes the 5' eukaryotic cap from mRNA transcripts in a critical step of many cellular RNA decay pathways. Dcp2 is a dynamic enzyme that functions in concert with the essential activator Dcp1 and a diverse set of coactivators to selectively and efficiently decap target mRNAs in the cell. Here we present a 2.84 Å crystal structure of K. lactis Dcp1-Dcp2 in complex with coactivators Edc1 and Edc3, and with substrate analog bound to the Dcp2 active site. Our structure shows how Dcp2 recognizes cap substrate in the catalytically active conformation of the enzyme, and how coactivator Edc1 forms a three-way interface that bridges the domains of Dcp2 to consolidate the active conformation. Kinetic data reveal Dcp2 has selectivity for the first transcribed nucleotide during the catalytic step. The heterotetrameric Edc1-Dcp1-Dcp2-Edc3 structure shows how coactivators Edc1 and Edc3 can act simultaneously to activate decapping catalysis.
Organizational Affiliation:
Department of Pharmaceutical Chemistry, University of California, San Francisco, San Francisco, CA, 94158, USA.