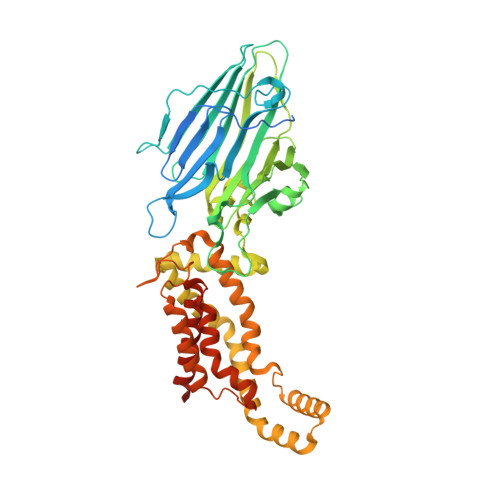
2.8-angstrom crystal structure of Escherichia coli YidC revealing all core regions, including flexible C2 loop.
Tanaka, Y., Izumioka, A., Abdul Hamid, A., Fujii, A., Haruyama, T., Furukawa, A., Tsukazaki, T.(2018) Biochem Biophys Res Commun 505: 141-145
- PubMed: 30241934
- DOI: https://doi.org/10.1016/j.bbrc.2018.09.043
- Primary Citation of Related Structures:
6AL2 - PubMed Abstract:
YidC/Alb3/Oxa1 family proteins are involved in the insertion and assembly of membrane proteins. The core five transmembrane regions of YidC, which are conserved in the protein family, form a positively charged cavity open to the cytoplasmic side. The cavity plays an important role in membrane protein insertion. In all reported structural studies of YidC, the second cytoplasmic loop (C2 loop) was disordered, limiting the understanding of its role. Here, we determined the crystal structure of YidC including the C2 loop at 2.8 Å resolution with R/R free = 21.8/27.5. This structure and subsequent molecular dynamics simulation indicated that the intrinsic flexible C2 loop covered the positively charged cavity. This crystal structure provides the coordinates of the complete core region including the C2 loop, which is valuable for further analyses of YidC.
Organizational Affiliation:
Graduate School of Science and Technology, Nara Institute of Science and Technology, Nara, 630-0192, Japan.