A High-Affinity Peptide Ligand Targeting Syntenin Inhibits Glioblastoma.
Haugaard-Kedstrom, L.M., Clemmensen, L.S., Sereikaite, V., Jin, Z., Fernandes, E.F.A., Wind, B., Abalde-Gil, F., Daberger, J., Vistrup-Parry, M., Aguilar-Morante, D., Leblanc, R., Egea-Jimenez, A.L., Albrigtsen, M., Jensen, K.E., Jensen, T.M.T., Ivarsson, Y., Vincentelli, R., Hamerlik, P., Andersen, J.H., Zimmermann, P., Lee, W., Stromgaard, K.(2021) J Med Chem 64: 1423-1434
- PubMed: 33502198
- DOI: https://doi.org/10.1021/acs.jmedchem.0c00382
- Primary Citation of Related Structures:
6AK2 - PubMed Abstract:
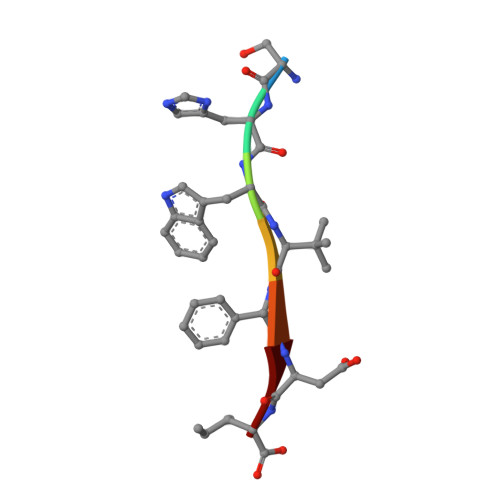
Despite the recent advances in cancer therapeutics, highly aggressive cancer forms, such as glioblastoma (GBM), still have very low survival rates. The intracellular scaffold protein syntenin, comprising two postsynaptic density protein-95/discs-large/zona occludens-1 (PDZ) domains, has emerged as a novel therapeutic target in highly malignant phenotypes including GBM. Here, we report the development of a novel, highly potent, and metabolically stable peptide inhibitor of syntenin, KSL-128114, which binds the PDZ1 domain of syntenin with nanomolar affinity. KSL-128114 is resistant toward degradation in human plasma and mouse hepatic microsomes and displays a global PDZ domain selectivity for syntenin. An X-ray crystal structure reveals that KSL-128114 interacts with syntenin PDZ1 in an extended noncanonical binding mode. Treatment with KSL-128114 shows an inhibitory effect on primary GBM cell viability and significantly extends survival time in a patient-derived xenograft mouse model. Thus, KSL-128114 is a novel promising candidate with therapeutic potential for highly aggressive tumors, such as GBM.
Organizational Affiliation:
Center for Biopharmaceuticals, Department of Drug Design and Pharmacology, University of Copenhagen, Universitetsparken 2, 2100 Copenhagen, Denmark.