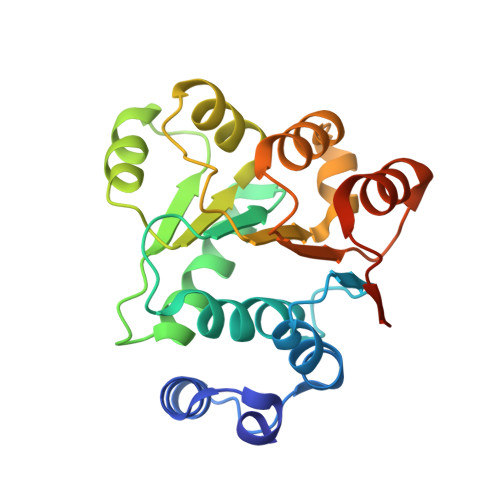
Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Chen, X., Wang, C., Zhang, X., Tian, T., Zang, J.(2018) Acta Crystallogr F Struct Biol Commun 74: 704-709
- PubMed: 30387775
- DOI: https://doi.org/10.1107/S2053230X1801292X
- Primary Citation of Related Structures:
6AIB, 6AIC - PubMed Abstract:
CshA is a DEAD-box RNA helicase that belongs to the DExD/H-box family of proteins, which generally have an RNA-dependent ATPase activity. In Staphylococcus aureus, CshA was identified as a component of the RNA degradosome and plays important roles in RNA turnover. In this study, the crystal structures of the N-terminal RecA-like domain 1 of S. aureus CshA (SaCshA R1 ) and of its complex with AMP (SaCshA R1 -AMP) are reported at resolutions of 1.5 and 1.8 Å, respectively. SaCshA R1 adopts a conserved α/β RecA-like structure with seven parallel strands surrounded by nine α-helices. The Q motif and motif I are responsible for the binding of the adenine group and phosphate group of AMP, respectively. Structure comparison of SaCshA R1 -AMP and SaCshA R1 reveals that motif I undergoes a conformational change upon AMP binding. Isothermal titration calorimetry assays further conformed the essential roles of Phe22 in the Q motif and Lys52 in motif I for binding ATP, indicating a conserved substrate-binding mechanism in SaCshA compared with other DEAD-box RNA helicases.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at Microscale and School of Life Sciences, Collaborative Innovation Center of Chemistry for Life Science, University of Science and Technology of China, 96 Jinzhai Road, Hefei, Anhui 230026, People's Republic of China.