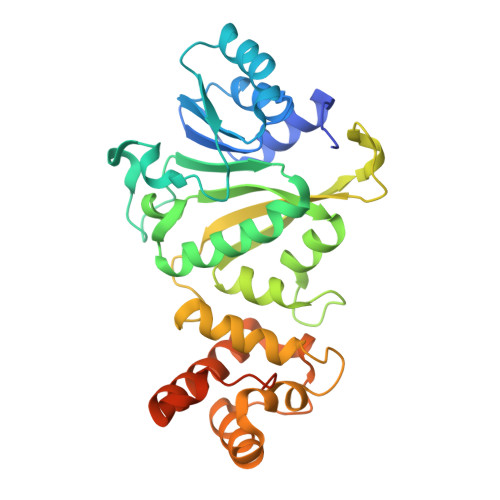
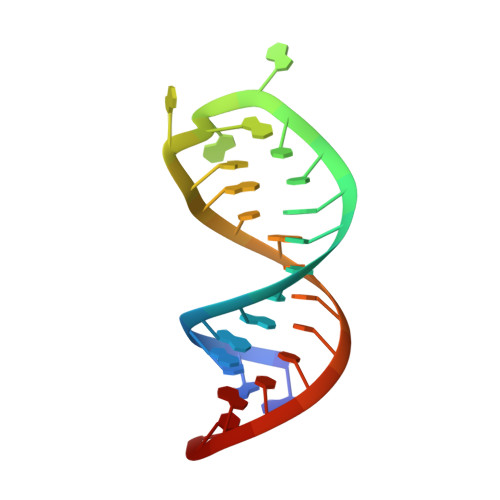
Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Liu, X., Shen, S., Wu, P., Li, F., Liu, X., Wang, C., Gong, Q., Wu, J., Yao, X., Zhang, H., Shi, Y.(2019) Nucleic Acids Res 47: 7648-7665
- PubMed: 31251801
- DOI: https://doi.org/10.1093/nar/gkz505
- Primary Citation of Related Structures:
6AAS, 6AAU, 6AAX, 6AJK - PubMed Abstract:
Mitochondria are essential molecular machinery for the maintenance of cellular energy supply by the oxidative phosphorylation system (OXPHOS). Mitochondrial transcription factor B1 (TFB1M) is a dimethyltransferase that maintains mitochondrial homeostasis by catalyzing dimethylation of two adjacent adenines located in helix45 (h45) of 12S rRNA. This m62A modification is indispensable for the assembly and maturation of human mitochondrial ribosomes. However, both the mechanism of TFB1M catalysis and the precise function of TFB1M in mitochondrial homeostasis are unknown. Here we report the crystal structures of a ternary complex of human (hs) TFB1M-h45-S-adenosyl-methionine and a binary complex hsTFB1M-h45. The structures revealed a distinct mode of hsTFB1M interaction with its rRNA substrate and with the initial enzymatic state involved in m62A modification. The suppression of hsTFB1M protein level or the overexpression of inactive hsTFB1M mutants resulted in decreased ATP production and reduced expression of components of the mitochondrial OXPHOS without affecting transcription of the corresponding genes and their localization to the mitochondria. Therefore, hsTFB1M regulated the translation of mitochondrial genes rather than their transcription via m62A modification in h45.
Organizational Affiliation:
School of Life Sciences, University of Science & Technology of China, Hefei 230027, China.