RNA 5-Methylcytosine Facilitates the Maternal-to-Zygotic Transition by Preventing Maternal mRNA Decay.
Yang, Y., Wang, L., Han, X., Yang, W.L., Zhang, M., Ma, H.L., Sun, B.F., Li, A., Xia, J., Chen, J., Heng, J., Wu, B., Chen, Y.S., Xu, J.W., Yang, X., Yao, H., Sun, J., Lyu, C., Wang, H.L., Huang, Y., Sun, Y.P., Zhao, Y.L., Meng, A., Ma, J., Liu, F., Yang, Y.G.(2019) Mol Cell 75: 1188-1202.e11
- PubMed: 31399345
- DOI: https://doi.org/10.1016/j.molcel.2019.06.033
- Primary Citation of Related Structures:
6A6J - PubMed Abstract:
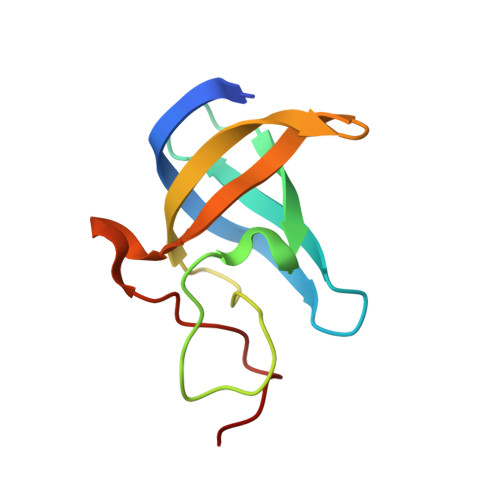
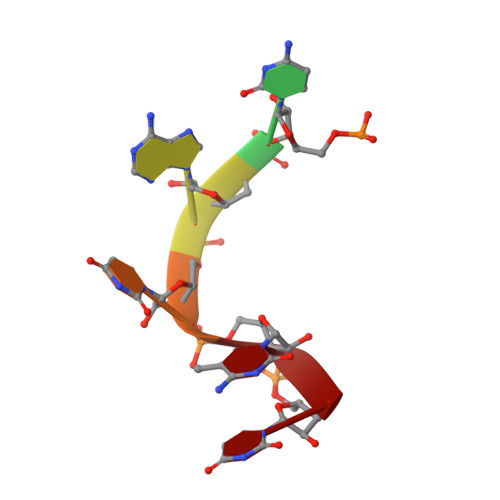
The maternal-to-zygotic transition (MZT) is a conserved and fundamental process during which the maternal environment is converted to an environment of embryonic-driven development through dramatic reprogramming. However, how maternally supplied transcripts are dynamically regulated during MZT remains largely unknown. Herein, through genome-wide profiling of RNA 5-methylcytosine (m 5 C) modification in zebrafish early embryos, we found that m 5 C-modified maternal mRNAs display higher stability than non-m 5 C-modified mRNAs during MZT. We discovered that Y-box binding protein 1 (Ybx1) preferentially recognizes m 5 C-modified mRNAs through π-π interactions with a key residue, Trp45, in Ybx1's cold shock domain (CSD), which plays essential roles in maternal mRNA stability and early embryogenesis of zebrafish. Together with the mRNA stabilizer Pabpc1a, Ybx1 promotes the stability of its target mRNAs in an m 5 C-dependent manner. Our study demonstrates an unexpected mechanism of RNA m 5 C-regulated maternal mRNA stabilization during zebrafish MZT, highlighting the critical role of m 5 C mRNA modification in early development.
Organizational Affiliation:
CAS Key Laboratory of Genomic and Precision Medicine, Collaborative Innovation Center of Genetics and Development, College of Future Technology, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China; Sino-Danish College, University of Chinese Academy of Sciences, Beijing 101408, China; Institute of Stem Cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China.