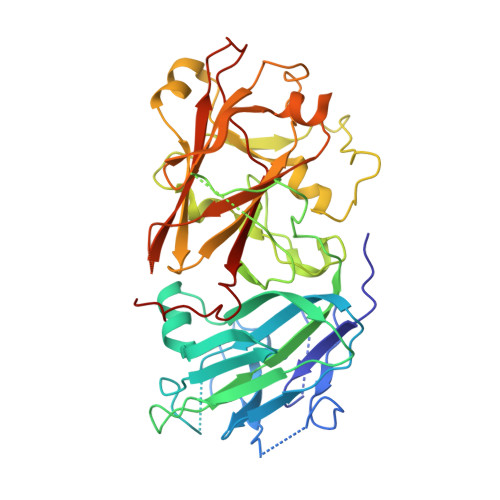
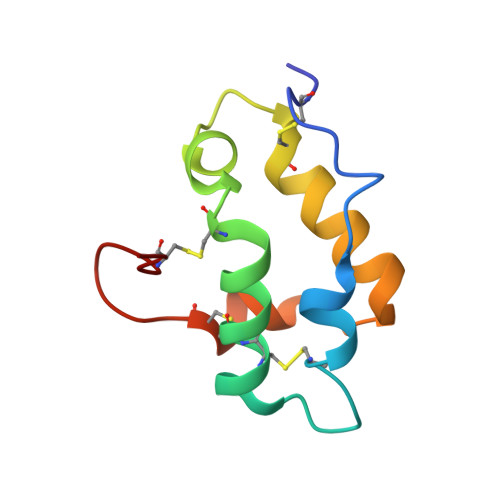
Mechanisms of RALF peptide perception by a heterotypic receptor complex.
Xiao, Y., Stegmann, M., Han, Z., DeFalco, T.A., Parys, K., Xu, L., Belkhadir, Y., Zipfel, C., Chai, J.(2019) Nature 572: 270-274
- PubMed: 31291642
- DOI: https://doi.org/10.1038/s41586-019-1409-7
- Primary Citation of Related Structures:
6A5A, 6A5B, 6A5C, 6A5D, 6A5E - PubMed Abstract:
Receptor kinases of the Catharanthus roseus RLK1-like (CrRLK1L) family have emerged as important regulators of plant reproduction, growth and responses to the environment 1 . Endogenous RAPID ALKALINIZATION FACTOR (RALF) peptides 2 have previously been proposed as ligands for several members of the CrRLK1L family 1 . However, the mechanistic basis of this perception is unknown. Here we report that RALF23 induces a complex between the CrRLK1L FERONIA (FER) and LORELEI (LRE)-LIKE GLYCOSYLPHOSPHATIDYLINOSITOL (GPI)-ANCHORED PROTEIN 1 (LLG1) to regulate immune signalling. Structural and biochemical data indicate that LLG1 (which is genetically important for RALF23 responses) and the related LLG2 directly bind RALF23 to nucleate the assembly of RALF23-LLG1-FER and RALF23-LLG2-FER heterocomplexes, respectively. A conserved N-terminal region of RALF23 is sufficient for the biochemical recognition of RALF23 by LLG1, LLG2 or LLG3, and binding assays suggest that other RALF peptides that share this conserved N-terminal region may be perceived by LLG proteins in a similar manner. Structural data also show that RALF23 recognition is governed by the conformationally flexible C-terminal sides of LLG1, LLG2 and LLG3. Our work reveals a mechanism of peptide perception in plants by GPI-anchored proteins that act together with a phylogenetically unrelated receptor kinase. This provides a molecular framework for understanding how diverse RALF peptides may regulate multiple processes, through perception by distinct heterocomplexes of CrRLK1L receptor kinases and GPI-anchored proteins of the LRE and LLG family.
Organizational Affiliation:
School of Life Sciences, Innovation Center for Structural Biology, Tsinghua-Peking Joint Center for Life Sciences, Tsinghua University, Beijing, China.