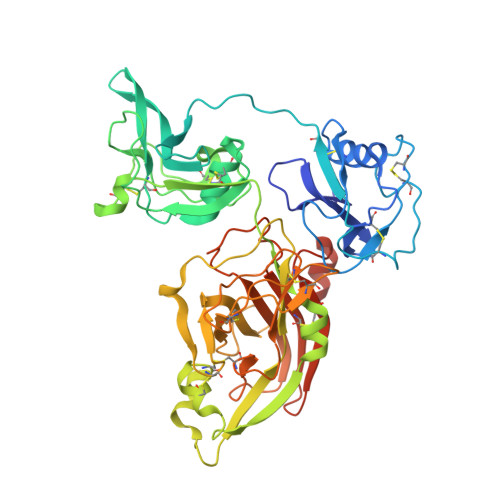
Crystal structure of human lysyl oxidase-like 2 (hLOXL2) in a precursor state.
Zhang, X., Wang, Q., Wu, J., Wang, J., Shi, Y., Liu, M.(2018) Proc Natl Acad Sci U S A 115: 3828-3833
- PubMed: 29581294
- DOI: https://doi.org/10.1073/pnas.1720859115
- Primary Citation of Related Structures:
5ZE3 - PubMed Abstract:
Lysyl oxidases (LOXs), a type of copper- and lysyl tyrosylquinone (LTQ) -dependent amine oxidase, catalyze the oxidative deamination of lysine residues of extracellular matrix (ECM) proteins such as elastins and collagens and generate aldehyde groups. The oxidative deamination of lysine represents the foundational step for the cross-linking of elastin and collagen and thus is crucial for ECM modeling. Despite their physiological significance, the structure of this important family of enzymes remains elusive. Here we report the crystal structure of human lysyl oxidase-like 2 (hLOXL2) at 2.4-Å resolution. Unexpectedly, the copper-binding site of hLOXL2 is occupied by zinc, which blocks LTQ generation and the enzymatic activity of hLOXL2 in our in vitro assay. Biochemical analysis confirms that copper loading robustly activates hLOXL2 and supports LTQ formation. Furthermore, the LTQ precursor residues in the structure are distanced by 16.6 Å, corroborating the notion that the present structure may represent a precursor state and that pronounced conformational rearrangements would be required for protein activation. The structure presented here establishes an important foundation for understanding the structure-function relationship of LOX proteins and will facilitate LOX-targeting drug discovery.
Organizational Affiliation:
Beijing Advanced Innovation Center for Structural Biology, Tsinghua University, 100084 Beijing, China.