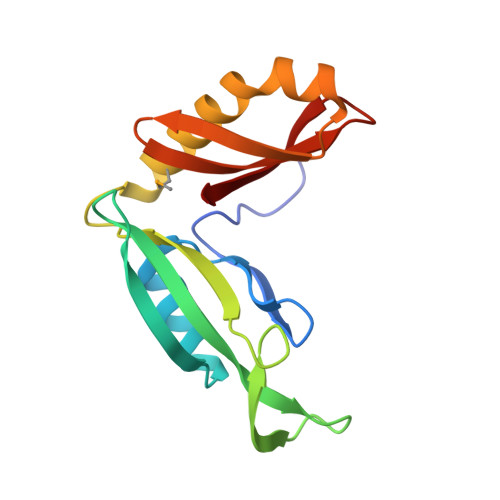
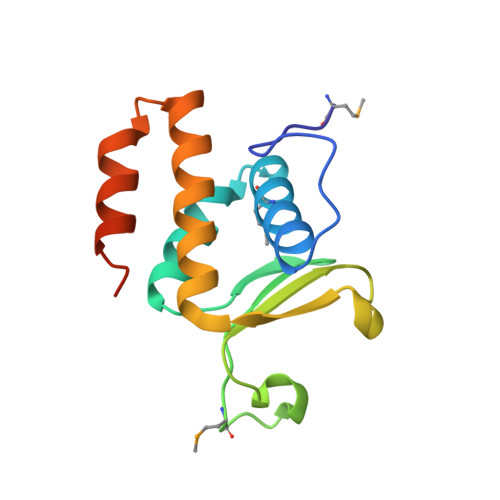
Functional plasticity of antibacterial EndoU toxins.
Michalska, K., Quan Nhan, D., Willett, J.L.E., Stols, L.M., Eschenfeldt, W.H., Jones, A.M., Nguyen, J.Y., Koskiniemi, S., Low, D.A., Goulding, C.W., Joachimiak, A., Hayes, C.S.(2018) Mol Microbiol 109: 509-527
- PubMed: 29923643
- DOI: https://doi.org/10.1111/mmi.14007
- Primary Citation of Related Structures:
5HKQ - PubMed Abstract:
Bacteria use several different secretion systems to deliver toxic EndoU ribonucleases into neighboring cells. Here, we present the first structure of a prokaryotic EndoU toxin in complex with its cognate immunity protein. The contact-dependent growth inhibition toxin CdiA-CT STECO31 from Escherichia coli STEC_O31 adopts the eukaryotic EndoU fold and shares greatest structural homology with the nuclease domain of coronavirus Nsp15. The toxin contains a canonical His-His-Lys catalytic triad in the same arrangement as eukaryotic EndoU domains, but lacks the uridylate-specific ribonuclease activity that characterizes the superfamily. Comparative sequence analysis indicates that bacterial EndoU domains segregate into at least three major clades based on structural variations in the N-terminal subdomain. Representative EndoU nucleases from clades I and II degrade tRNA molecules with little specificity. In contrast, CdiA-CT STECO31 and other clade III toxins are specific anticodon nucleases that cleave tRNA Glu between nucleotides C37 and m 2 A38. These findings suggest that the EndoU fold is a versatile scaffold for the evolution of novel substrate specificities. Such functional plasticity may account for the widespread use of EndoU effectors by diverse inter-bacterial toxin delivery systems.
Organizational Affiliation:
Midwest Center for Structural Genomics, Argonne National Laboratory, Argonne, IL, USA.