Crystal structure determination of Cys2Ala mutant of Bile Salt Hydrolase from Enterococcus feacalis
Ramasamy, S., Deepak, C., Varshney, N.K., Suresh, C.G.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Choloylglycine hydrolase | 323 | Enterococcus faecalis | Mutation(s): 1 Gene Names: ef0040 EC: 3.5.1.24 | 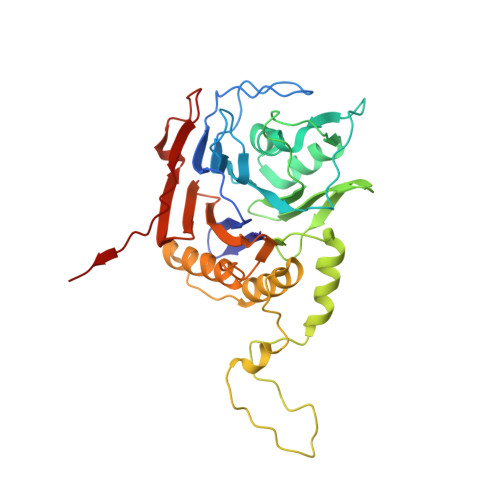 | |
UniProt | |||||
Find proteins for Q8KUA7 (Enterococcus faecalis) Explore Q8KUA7 Go to UniProtKB: Q8KUA7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8KUA7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 91.187 | α = 90 |
| b = 91.187 | β = 90 |
| c = 156.505 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| CSIR network project HUM | India | BSC0119 |