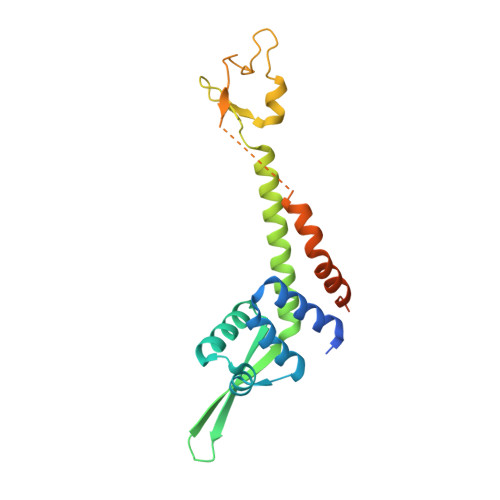
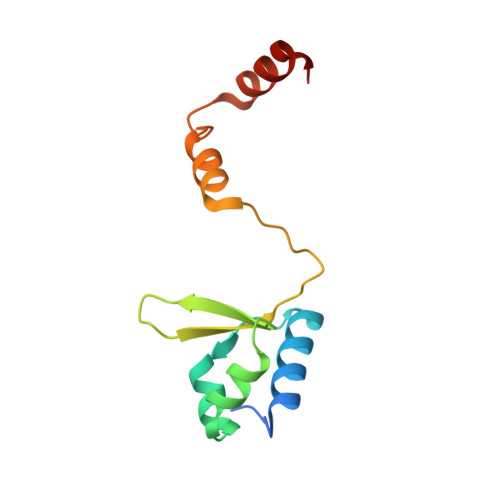
Crystal Structure of Human General Transcription Factor TFIIE at Atomic Resolution
Miwa, K., Kojima, R., Obita, T., Ohkuma, Y., Tamura, Y., Mizuguchi, M.(2016) J Mol Biol 428: 4258-4266
- PubMed: 27639436
- DOI: https://doi.org/10.1016/j.jmb.2016.09.008
- Primary Citation of Related Structures:
5GPY - PubMed Abstract:
In eukaryotes, RNA polymerase II requires general transcription factors to initiate mRNA transcription. TFIIE subunits α and β form a heterodimer and recruit TFIIH to complete the assembly of the pre-initiation complex. Here, we have determined the crystal structure of human TFIIE at atomic resolution. The N-terminal half of TFIIEα forms an extended winged helix (WH) domain with an additional helix, followed by a zinc-finger domain. TFIIEβ contains the WH2 domain, followed by two coiled-coil helices intertwining with TFIIEα. We also showed that TFIIEα binds to TFIIEβ with nanomolar affinity using isothermal titration calorimetry. In addition, mutations on the residues involved in the interactions resulted in severe growth defects in yeast. Lack of the C-terminal region of yeast TFIIEβ causes a mild growth defect in vivo. These findings provide a structural basis for understanding the functional mechanisms of TFIIE in the context of pre-initiation complex formation and transcription initiation.
Organizational Affiliation:
Faculty of Pharmaceutical Sciences, University of Toyama, 2630 Sugitani, Toyama 930-0194, Japan.