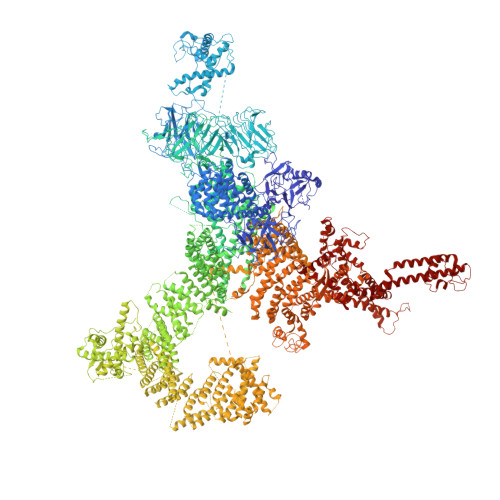
The Central domain of RyR1 is the transducer for long-range allosteric gating of channel opening
Bai, X.C., Yan, Z., Wu, J.P., Li, Z.Q., Yan, N.(2016) Cell Res 26: 995-1006
- PubMed: 27468892
- DOI: https://doi.org/10.1038/cr.2016.89
- Primary Citation of Related Structures:
5GKY, 5GKZ, 5GL0, 5GL1 - PubMed Abstract:
The ryanodine receptors (RyRs) are intracellular calcium channels responsible for rapid release of Ca(2+) from the sarcoplasmic/endoplasmic reticulum (SR/ER) to the cytoplasm, which is essential for the excitation-contraction (E-C) coupling of cardiac and skeletal muscles. The near-atomic resolution structure of closed RyR1 revealed the molecular details of this colossal channel, while the long-range allosteric gating mechanism awaits elucidation. Here, we report the cryo-EM structures of rabbit RyR1 in three closed conformations at about 4 Å resolution and an open state at 5.7 Å. Comparison of the closed RyR1 structures shows a breathing motion of the cytoplasmic platform, while the channel domain and its contiguous Central domain remain nearly unchanged. Comparison of the open and closed structures shows a dilation of the S6 tetrahelical bundle at the cytoplasmic gate that leads to channel opening. During the pore opening, the cytoplasmic "O-ring" motif of the channel domain and the U-motif of the Central domain exhibit coupled motion, while the Central domain undergoes domain-wise displacement. These structural analyses provide important insight into the E-C coupling in skeletal muscles and identify the Central domain as the transducer that couples the conformational changes of the cytoplasmic platform to the gating of the central pore.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Cambridge Biomedical Campus, Cambridge CB2 0QH, UK.