The Vaccinia Virus H3 Envelope Protein, a Major Target of Neutralizing Antibodies, Exhibits a Glycosyltransferase Fold and Binds UDP-Glucose.
Singh, K., Gittis, A.G., Gitti, R.K., Ostazeski, S.A., Su, H.P., Garboczi, D.N.(2016) J Virol 90: 5020-5030
- PubMed: 26937025
- DOI: https://doi.org/10.1128/JVI.02933-15
- Primary Citation of Related Structures:
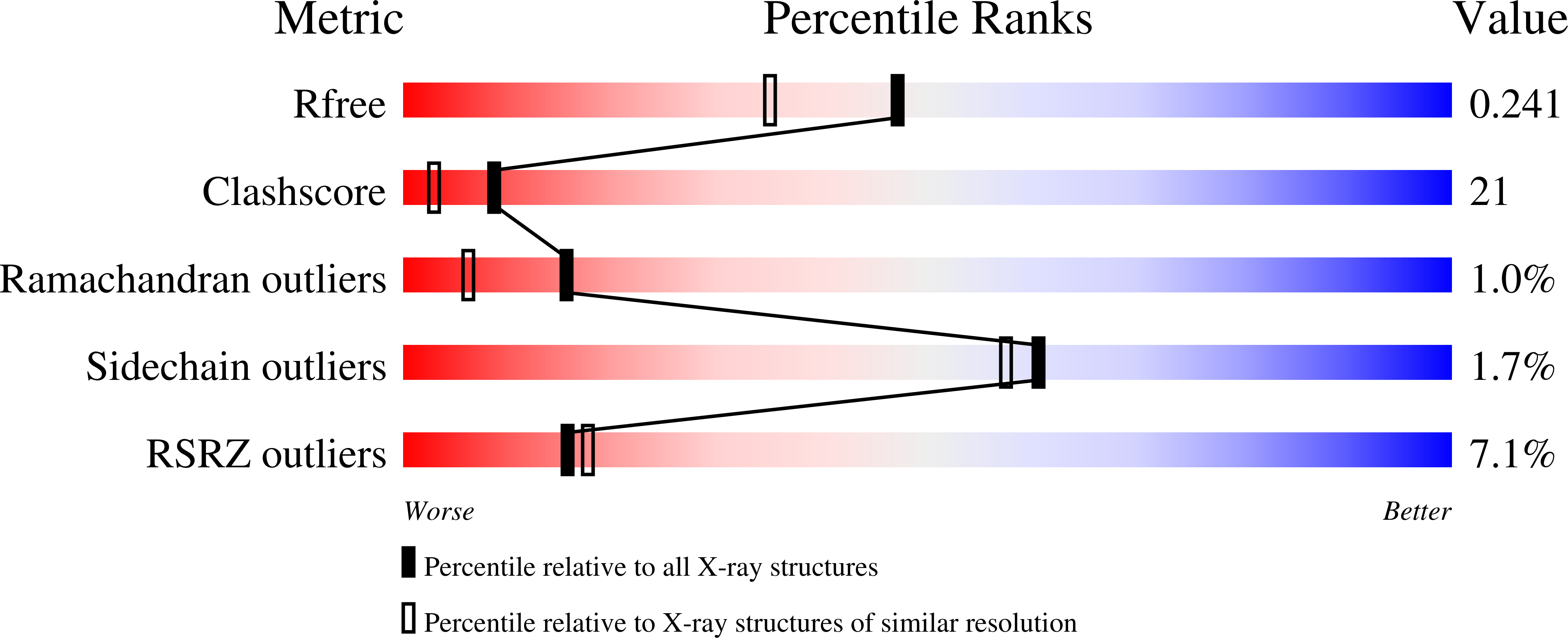
5EJ0 - PubMed Abstract:
The highly conserved H3 poxvirus protein is a major target of the human antibody response against poxviruses and is likely a key contributor to protection against infection. Here, we present the crystal structure of H3 from vaccinia virus at a 1.9-Å resolution. H3 looks like a glycosyltransferase, a family of enzymes that transfer carbohydrate molecules to a variety of acceptor substrates. Like glycosyltransferases, H3 binds UDP-glucose, as shown by saturation transfer difference (STD) nuclear magnetic resonance (NMR) spectroscopy, and this binding requires Mg(2+) Mutation of the glycosyltransferase-like metal ion binding motif in H3 greatly diminished its binding to UDP-glucose. We found by flow cytometry that H3 binds to the surface of human cells but does not bind well to cells that are deficient in surface glycosaminoglycans. STD NMR experiments using a heparin sulfate decasaccharide confirmed that H3 binds heparin sulfate. We propose that a surface of H3 with an excess positive charge may be the binding site for heparin. Heparin binding and glycosyltransferase activity may be involved in the function of H3 in the poxvirus life cycle.
Organizational Affiliation:
Structural Biology Section, Research Technologies Branch, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, Maryland, USA ksingh@niaid.nih.gov dgarboczi@niaid.nih.gov.