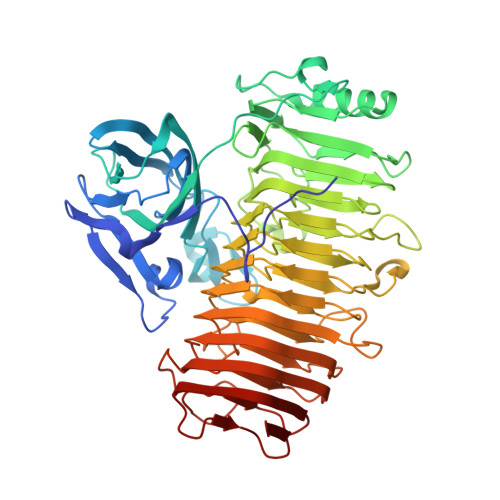
Crystal structure of the catalytic unit of GH 87-type alpha-1,3-glucanase Agl-KA from Bacillus circulans.
Yano, S., Suyotha, W., Oguro, N., Matsui, T., Shiga, S., Itoh, T., Hibi, T., Tanaka, Y., Wakayama, M., Makabe, K.(2019) Sci Rep 9: 15295-15295
- PubMed: 31653959
- DOI: https://doi.org/10.1038/s41598-019-51822-5
- Primary Citation of Related Structures:
5ZRU - PubMed Abstract:
Glycoside hydrolase (GH) 87-type α-1,3-glucanase hydrolyses the α-1,3-glucoside linkages of α-1,3-glucan, which is found in fungal cell walls and extracellular polysaccharides produced by oral Streptococci. In this study, we report on the molecular structure of the catalytic unit of GH 87-type α-1,3-glucanase, Agl-KA, from Bacillus circulans, as determined by x-ray crystallography at a resolution of 1.82 Å. The catalytic unit constitutes a complex structure of two tandemly connected domains-the N-terminal galactose-binding-like domain and the C-terminal right-handed β-helix domain. While the β-helix domain is widely found among polysaccharide-processing enzymes, complex formation with the galactose-binding-like domain was observed for the first time. Biochemical assays showed that Asp1067, Asp1090 and Asp1091 are important for catalysis, and these residues are indeed located at the putative substrate-binding cleft, which forms a closed end and explains the product specificity.
Organizational Affiliation:
Department of Biochemical Engineering, Graduate School of Sciences and Engineering, Yamagata University, Jonan, Yonezawa, Yamagata, 992-8510, Japan.