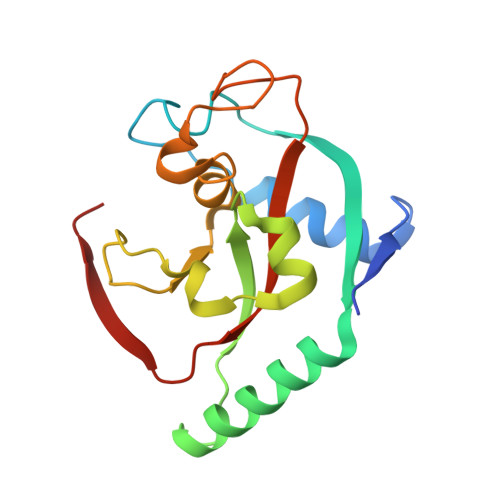
Discovery of Novel Spiroindoline Derivatives as Selective Tankyrase Inhibitors.
Shirai, F., Tsumura, T., Yashiroda, Y., Yuki, H., Niwa, H., Sato, S., Chikada, T., Koda, Y., Washizuka, K., Yoshimoto, N., Abe, M., Onuki, T., Mazaki, Y., Hirama, C., Fukami, T., Watanabe, H., Honma, T., Umehara, T., Shirouzu, M., Okue, M., Kano, Y., Watanabe, T., Kitamura, K., Shitara, E., Muramatsu, Y., Yoshida, H., Mizutani, A., Seimiya, H., Yoshida, M., Koyama, H.(2019) J Med Chem 62: 3407-3427
- PubMed: 30883102
- DOI: https://doi.org/10.1021/acs.jmedchem.8b01888
- Primary Citation of Related Structures:
5ZQO, 5ZQP, 5ZQQ, 5ZQR, 6A84 - PubMed Abstract:
The canonical WNT pathway plays an important role in cancer pathogenesis. Inhibition of poly(ADP-ribose) polymerase catalytic activity of the tankyrases (TNKS/TNKS2) has been reported to reduce the Wnt/β-catenin signal by preventing poly ADP-ribosylation-dependent degradation of AXIN, a negative regulator of Wnt/β-catenin signaling. With the goal of investigating the effects of tankyrase and Wnt pathway inhibition on tumor growth, we set out to find small-molecule inhibitors of TNKS/TNKS2 with suitable drug-like properties. Starting from 1a, a high-throughput screening hit, the spiroindoline derivative 40c (RK-287107) was discovered as a potent TNKS/TNKS2 inhibitor with >7000-fold selectivity against the PARP1 enzyme, which inhibits WNT-responsive TCF reporter activity and proliferation of human colorectal cancer cell line COLO-320DM. RK-287107 also demonstrated dose-dependent tumor growth inhibition in a mouse xenograft model. These observations suggest that RK-287107 is a promising lead compound for the development of novel tankyrase inhibitors as anticancer agents.
Organizational Affiliation:
RIKEN Program for Drug Discovery and Medical Technology Platforms , 2-1 Hirosawa , Wako , Saitama 351-0198 , Japan.